The nuclear to cytoplasmic ratio directly regulates zygotic transcription in Drosophila
Posted on: 14 October 2019
Preprint posted on 13 September 2019
Article now published in Proceedings of the National Academy of Sciences at http://dx.doi.org/10.1073/pnas.2010210118
Switching it up: the nuclear-cytoplasmic ratio directly regulates transcription of a switch-like gene, while indirectly affecting other zygotic genes.
Selected by Jessica XieCategories: developmental biology
Background
Early embryo development in many species begins with a cleavage stage, in which the single-celled zygote undergoes an iconic series of rapid, synchronous cell divisions without much mass increase or transcriptional activity. The end of this cleavage phase—the midblastula transition (MBT)—is associated with two other phenomena: the cell cycle slows, and zygotic genome activation (ZGA) begins.
The MBT, with its attendant changes in cell cycle length and transcription, is well-established to be controlled by the ratio of nuclear (i.e. DNA) to cytoplasmic material, or “N/C ratio”—for example, removal of cytoplasm accelerates MBT timing, while haploid embryos have delayed MBT. However, as cell cycle length and ZGA are themselves also highly interdependent (for example, prolonging the cell cycle with cell cycle inhibitors can induce premature ZGA), it has proven challenging to disentangle how exactly the N/C ratio affects either of these processes individually.
Such an investigation would require both high temporal resolution (to distinguish initial changes from secondary effects) and spatial information (as many early genes have specific spatial patterning). Moreover, ploidy manipulations—a common method of increasing DNA amounts in vivo—add the additional confounding factor of increased template availability.
A previous report by these authors (Amodeo et al., 2015) provided an ingenious demonstration of N/C-ratio-dependent transcription above the threshold of approximately 50 ng DNA/µL cytoplasm. However, this study had not identified which gene transcript(s) specifically show N/C-ratio-dependence; it was also an in vitro assay controlling N/C ratio by adding purified sperm chromatin to cytosolic extracts of unfertilized Xenopus eggs.
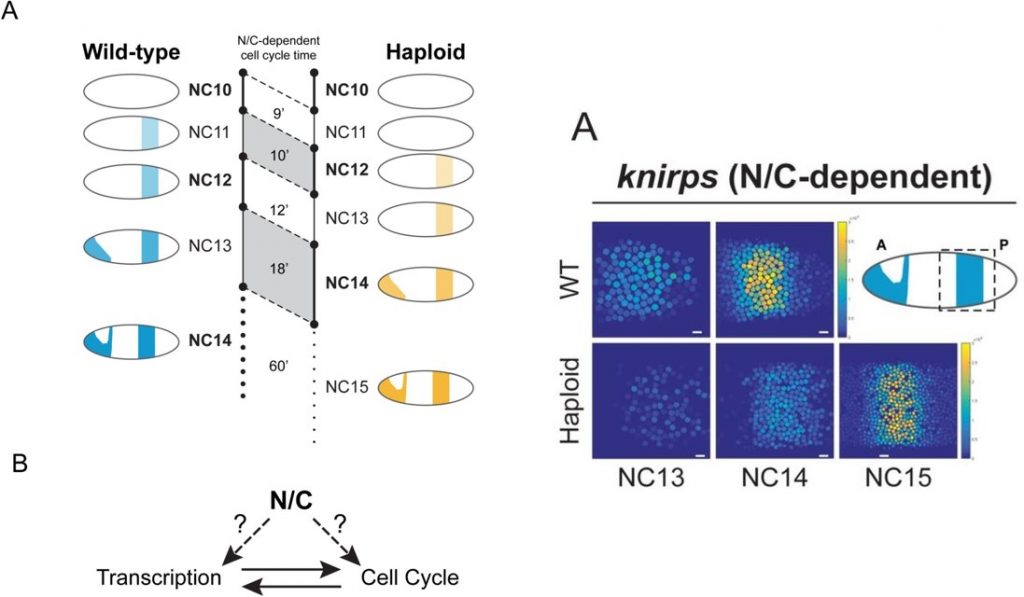
In this study, the authors address many of the aforementioned technical limitations by using the fluorescence-based MS2-MCP system in Drosophila to monitor the transcription of several candidate genes in vivo. This system first involves the insertion of multiple MS2 repeat DNA sequences into the gene of interest. When transcribed, the MS2 sequences form mRNA stem loops, which are bound by the MCP-GFP protein to activate fluorescence locally, rapidly (with <30 s temporal resolution), and in a manner proportional to transcription levels.
In order to distinguish effects of N/C ratio and cell cycle length on transcription, they performed measurements in several genetic mutants: WT diploids, haploid mutants (which have reduced N/C ratio—but, potentially confoundingly, also shorter cell cycles), and short-cell-cycle diploid mutants (to isolate effects of short cell cycle duration). This study particularly focused on characterizing the transcription of some genes previously found to have N/C-ratio-dependent expression, and some that did not—such as knirps (kni) and snail (sna), respectively.
Results
The first findings were unexpected: regardless of their prior classification, all studied genes were found to have similar transcriptional deficits in haploid mutants. Transcription of the N/C-independent gene sna and the N/C-ratio-dependent genes kni and gt were all delayed by one cell division cycle in haploid embryos relative to WT diploids, suggesting that the N/C ratio may have a greater effect on transcription than previously anticipated.
The authors note that maximum fluorescence amplitude (maximum transcriptional activity per cell) of several genes was reduced in haploids compared to WT, but they attribute this to the shorter cell cycle and consequent early termination of transcription in haploids—transcriptional activity never reaches its steady state maxima. In support of this interpretation, they find no difference in maximum transcriptional activity between haploids and short cell cycle diploid mutants. They thus conclude that the N/C ratio affects cell cycle duration and thereby has an indirect effect on the transcriptional output of all genes.
After finding that all tested genes showed no difference in transcriptional output per nucleus between haploids and WT, the authors looked at probability of transcriptional activation, and found that the proportion of nuclei actively transcribing each gene also remained constant throughout cell division cycles 11–15 in both haploids and WT.
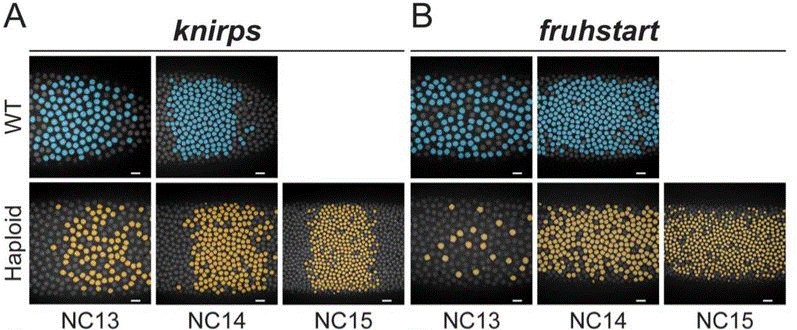
One exception stood out, however. Unlike other genes tested, the cell cycle regulator frs displayed rapid switchlike activation at a very specific stage: it was expressed by 16% of WT nuclei in cycle 12, then 80% in the subsequent cycle 13. This increase was delayed by one cell cycle in haploids, occurring between cycles 13 & 14. Crucially, however, this occurred at the normal time (cycles 12–13) in short-cell-cycle diploid mutants, indicating that the delay in activation was not due to cell cycle duration defects, and supporting the authors’ conclusion that frs activation is regulated by ploidy—i.e. directly by the N/C ratio.
Questions for the authors:
- What proportion of genes would you speculate to be directly N/C-ratio-dependent?
- What components of frs transcription regulation might you expect to be involved in N/C ratio sensing?
- Have you tried any other methods of changing N/C ratio? The triumvirate of factors (N/C ratio, cell cycle length, transcription activation) are all so interdependent that it seems like it would be hard to conclude from just one perturbation that N/C ratio is the reason for the delay of frs activation in haploids!
- Do you expect that switchlike genes (similar to frs) would be likely to be N/C-ratio-dependent? In other words, would a good way to find directly N/C-ratio-dependent genes be to look for zygotic genes that show such transcriptional profiles?
References
Amodeo, A.A., Jukam, D., Straight, A.F., & Skotheim, J.M. (2015). Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proceedings of the National Academy of Sciences of the United States of America, 112(10), E1086-95. https://doi.org/10.1073/pnas.1413990112
doi: https://doi.org/10.1242/prelights.14492
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Ectopic head regeneration after nervous system ablation in a sea anemone
Isabella Cisneros
Hyaluronic Acid and Emergent Tissue Mechanics Orchestrate Digit Tip Regeneration
Jonathan Townson
Visually-guided compensation of deafening-induced song deterioration
Maitri Manjunath
preLists in the developmental biology category:
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Jonathan Townson, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)