Trophectoderm Potency is Retained Exclusively in Human Naïve Cells
Posted on: 1 March 2020 , updated on: 2 March 2020
Preprint posted on 4 February 2020
Article now published in Cell Stem Cell at http://dx.doi.org/10.1016/j.stem.2021.02.025
Regretting lineage decisions: Human naïve cells and the inner cell mass from human blastocysts retain the potential to generate trophectoderm-like cells.
Selected by Sergio MencheroCategories: developmental biology
Background & Summary:
Embryonic development is understood as a series of branching decisions taken under the control of lineage programs. These lineage bifurcations are generally unidirectional, and once a cell is committed to a lineage it cannot come back.
In mammals, after the first fate decision, cells are segregated in two lineages: trophectoderm (TE, future placenta) and inner cell mass (ICM, pluripotent population that will give rise to the embryo). In the mouse, during the specification of these first lineages there is a certain degree of plasticity (Posfai et al. 2017) but once they are established, cells from one lineage cannot contribute to the other lineage. Mouse trophoblast stem cells (mTSCs) and mouse embryonic stem cells (mESCs), the in vitro counterparts of TE and ICM respectively, retain the potential from the lineage they were derived from, although they can be interconverted to other lineages by genetic manipulations and modification of culture conditions (Garg et al. 2016).
Interestingly, naïve human ESCs (hESCs) have a transcriptional signature more similar to human morula cells, an earlier stage, than to the segregated ICM (Theunissen et al. 2016). Because at this stage the first lineages have not been segregated yet (Petropoulos et al. 2016), the putative potential of these naïve hESCs to contribute to TE fate has been a standing question in the field.
Major findings in the preprint:
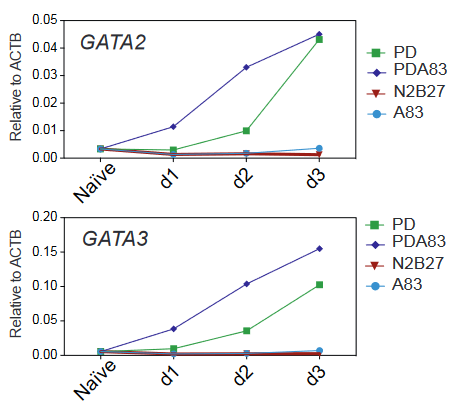
- Naïve human ESCs can upregulate TE-related markers. Naïve mESCs are cultured in the presence of a MEK inhibitor (PD) and maintain their capacity to self-renew. When naïve hESCs were cultured with PD alone, some of them differentiated and started to express TE-markers such as GATA2 and GATA3. The authors generated a GATA3 knock-in reporter and confirmed the upregulation of the reporter upon culture in PD.
- Inhibition of Nodal and Activin pathway increases TE differentiation potential. When adding A83, an inhibitor of the Nodal and Activin pathways, to the culture media in addition to PD, the GATA3 reporter and other TE markers were activated earlier and stronger. Global transcriptomic analysis of naïve hESCs with PD+A83 showed that cells progressively acquired a TE signature similar to the TE of human embryos.
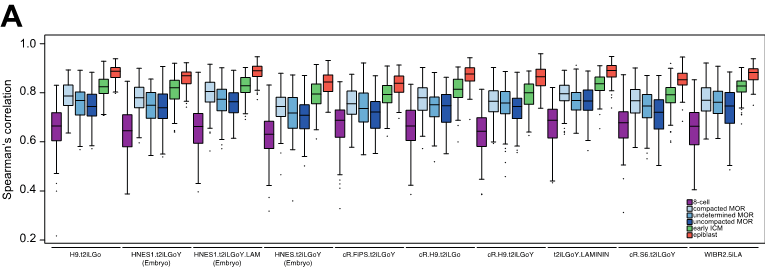
- Human ESCs with different transcriptomic signatures including primed hESCs, extended-potential hESCs (hEPSCs) and naïve hESCs behave differently upon inhibition of MEK and Nodal. It had been reported that primed hESCs and hEPSCs could differentiate toward TE in a BMP-dependent manner. However, in this preprint, the authors show that primed hESCs and hEPSCs acquire an amnion-like transcriptome upon culture with PD+A83 inhibitors instead of TE identity as naïve hESCs do. Moreover, naïve hESCs follow a BMP-independent differentiation path to differentiate to TE. This suggests that not all the human ESCs have the same default differentiation program, and differences in potentiality could affect the differentiation trajectories.
- Perturbations in key factors of the pluripotency and TE networks affect the potential of naïve hESCs to differentiate to TE.
- Targeted mutagenesis of the pluripotency-associated transcription factors OCT4, SOX2 and NANOG using CRISPR/Cas9 led to upregulation of TE markers, even when cells were cultured on N2B27 which does not promote TE differentiation in control conditions.
- Forced expression of NANOG prevented the upregulation of TE markers.
- TFAP2C and YAP mutant hESCs showed a reduced induction of TE markers as compared to control cells suggesting an important role for TE differentiation.
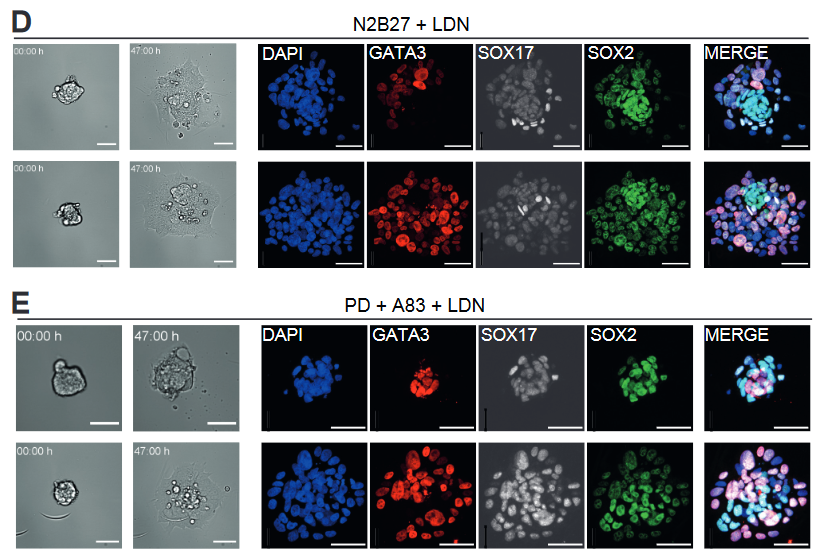
- ICM from human embryos retain the potential to generate TE cells. Culture of isolated ICMs in N2B27 medium or in PD+A83 gave rise to flattened cells that expressed GATA3. Those cells surrounded a cluster of SOX2 positive cells that represent the epiblast. In those outgrowth cultures, some cells with a primitive endoderm identity, recognised by the expression of SOX17, were also present. In vivo, cells from the ICM are expected to segregate into SOX2 or SOX17 positive cells when the epiblast and the primitive endoderm are specified. However, the fact that cells with TE markers also emerge from ICM cells indicates that the human embryo retains a high degree of plasticity.
General comments
The scarce availability of human embryos, together with ethical issues that their usage can raise, make their in vitro counterparts a very valuable tool to understand human embryo development. The authors of this preprint use naïve hESCs to nicely demonstrate that they retain the ability to upregulate the TE transcriptional program under certain culture conditions. These findings highlight the potential of naïve hESCs and how they could help to understand the specification of the TE identity in future studies, including the activation or inactivation of specific regulatory elements.
One important aspect to investigate is whether naïve hESCs de-differentiate from an epiblast-like state or if naïve hESCs mimic a cell state prior to the first lineage decision and differences in culture conditions bias their progression to a TE-like or ICM-like identity.
Questions to the authors
- In your experiments, you see that naïve hESCs pass through an ICM-like cell population before their differentiation to TE. If naïve hESCs have a transcriptional signature more similar to human morula cells than to the segregated ICM as it has been proposed (Theunissen et al. 2016), why do think they need to pass through an ICM-like state? If they represent a stage prior to the first lineage decision, a priori it could be independent of the specification of the ICM.
- Have you evaluated how far can naïve hESCs differentiate into TE derivatives? Do you think the “capacitation” process can affect their potential to give rise to more differentiated tissues?
- Analysis of scRNA-seq data from human preimplantation embryos revealed that blastomeres are transcriptionally very similar until the early blastocyst stage (Petropoulos et al. 2016). Do you think that the fact that human ICM retains TE lineage plasticity is because the lineages have been segregated more recently than in the mouse? Do you see any differences when using ICMs from early or very late blastocysts? Would you expect this plasticity to occur in other species where the TE lineage is also specified late like the pig (Ramos-Ibeas et al. 2019)?
References
Garg et al. (2016) Capturing Identity and Fate Ex Vivo: Stem Cells From the Mouse Blastocyst. Curr Top Dev Biol 120, 361-400.
Petropoulos et al. (2016) Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 165(4), 1012-26
Posfai et al. (2017) Position- And Hippo Signaling-Dependent Plasticity During Lineage Segregation in the Early Mouse Embryo. Elife 6:e22906.
Ramos-Ibeas et al. (2019) Pluripotency and X Chromosome Dynamics Revealed in Pig Pre-Gastrulating Embryos by Single Cell Analysis. Nat Commun 10(1), 500
Theunissen et al. (2016) Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 19(4), 502-515.
doi: https://doi.org/10.1242/prelights.17446
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)