Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs
Preprint posted on 15 May 2018 https://www.biorxiv.org/content/early/2018/05/15/323105
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-019-09985-2
A recent preprint highlights an advance in CRISPR/Cas9 technology that would enable the independent regulation of different target genes in the same cell.
Selected by Samantha SeahCategories: molecular biology, synthetic biology
Summary
Utilising ligands to activate or deactivate sgRNAs provides a new way to regulate CRISPR-Cas9 activity in a dose-dependent manner, enabling manipulations to be carried out on different targets within the same cell.
Background
The CRISPR/Cas9 system almost needs no introduction – its ease of use has made it the most attractive system for genetic engineering today. The Cas9 nuclease is targeted to specific sites in the genome by the introduction of single guide RNAs (sgRNAs) complementary to the site of interest, enabling precise DNA cleavage that can be utilised to generate gene knockouts or knock-ins. Alternatively, the inactivation of both Cas9 nuclease domains and coupling to an enzymatic domain, such as to an activation domain or epigenetic modifier, enables the specific targeting of enzymatic activities to exact genomic coordinates. Similarly, the inactivated Cas9 protein (dCas9) is able to sterically repress transcription. Previous adaptations made to control CRISPR/Cas9 activity largely target Cas9 activity using chemical or optical inputs, which modulates all gene targets in the same direction. It is also possible to use multiple orthogonal CRISPR-Cas9 systems to modulate different targets in different directions. A key limitation of the CRISPR/Cas technology is the potential for off-target effects, which may introduce unwanted effects and limits the use of CRISPR-Cas9 in therapeutics.
Key Findings
The authors demonstrate the use of ligands to activate or deactivate sgRNA, and thus CRISPR/Cas9. This was carried out by the addition of ligand-binding aptamers to the sgRNAs utilised. By choosing to target sgRNAs, which vary between different targets, different manipulations can be carried out on different targets within the same cell.
Here, the authors chose to use the well-characterised small molecule, theophylline, and its corresponding aptamer, to establish the system. They tested three different strategies to link the aptamer to sgRNA, and eventually found that strand displacement, using sequences that enable alternative base pairing in the ligand-bound and ligand-free states, was most successful strategy. Multiple designs were responsive to theophylline in vitro, and these included designs that were activated or deactivated by the addition of theophylline. Utilising CRISPR-Cas9-mediated repression of fluorescent protein expression, they showed that the ligand-activated and inactivated sgRNAs had up to 11x and 13x dynamic ranges respectively, spanning over 50% of the range achieved by the controls. These sgRNAs also responded to changing concentrations of theophylline in a dose-dependent manner, suggesting that these sgRNAs could be used to precisely modulate gene expression.
The authors even utilised a second, related ligand-aptamer pair. The 3-methylxanthine (3mx) aptamer differs from that of theophylline by a single base-pair, and even as the theophylline aptamer is recognised by both ligands, the 3-methylxanthine aptamer only responds to its ligand, 3mx. They were then able to establish a system with two different fluorescent proteins under the control of different ligand-aptamer pairs, and demonstrated that fluorescent protein expression can be logically and temporally regulated by sequential addition and removal of the two ligands.
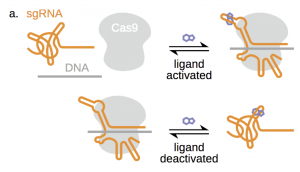
Figure 1a from the preprint: Principles of the technology. Small molecule aptamers are inserted into sgRNAs, resulting in the stabilisation of functional CRISPR-Cas9-sgRNA complexes either in the presence (above) or absence (below) of the ligand.
What I like about this work
I find the principles of this modification of CRISPR/Cas9 technology extremely elegant. By regulating individual sgRNAs, target genes can be independently modulated. The fact that they found both ligand activated and deactivated sgRNAs suggests that different genes can be regulated in opposite directions with a single ligand.
Outlook
The authors have established this system in bacteria cells, and it would be interesting to see if similar systems could be adapted for use in mammalian cells. The authors attempted this in both yeast and mammalian systems, and it did not appear to convincingly work in either. In addition, higher concentrations of theophylline was toxic to mammalian cells. It appears that alternative ligand-aptamer systems might be required to transfer this technology into eukaryotic cells.
(To the authors: Do you have any idea why the system did not appear to work in the eukaryotic cells tested? Do you have any idea how you might overcome this, to transfer the technology into eukaryotic cells?)
In addition, the availability of a variety of ligand-aptamer pairs that can act independently of each other would be ideal. Scientists could then pick and test ligand-aptamer pairs that suit their purposes.
Off-target effects are often problematic in CRISPR/Cas9 applications, and one way to minimise off-target effects is to only have Cas9 expressed for extremely short time periods. It would be interesting to see if this technology could be coupled with transient transfection to further minimise off-target effects.
Posted on: 14 June 2018
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
Also in the synthetic biology category:
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Genetically encoded multimeric tags for intracellular protein localisation in cryo-EM
Dissecting aneuploidy phenotypes by constructing Sc2.0 chromosome VII and SCRaMbLEing synthetic disomic yeast
preLists in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)