Impaired 26S proteasome causes learning and memory deficiency and induces neuroinflammation mediated by NF-κB in mice
Posted on: 22 August 2024 , updated on: 23 August 2024
Preprint posted on 7 April 2024
This preprint shows for the first time the importance of the Psmc1 gene as a regulator of proteasome activity, learning, memory and neuroinflammation (common Alzheimer’s disease symptoms).
Selected by Gustavo Stelzer, Marcus OliveiraCategories: animal behavior and cognition, biochemistry, neuroscience
Background
The ubiquitin-proteasome system (UPS) is a pathway commonly associated with protein degradation in eukaryotic cells(1,2). Selected proteins are tagged with polyubiquitin chains and, once marked, these proteins will be submitted to the catalytic activities of the proteasome(1,2). The proteasome is a multimeric protein complex which consists of specialized portions. The 20S proteasome consists of two outer α-rings and two inner β-rings, which serve as a gateway to protein entry and allows proteolytic activity, respectively(1,2). The 19S proteasome is an outer regulatory portion of the proteasome which has several functions, such as protein unfolding and α-ring opening to allow the protein to properly enter the proteolytic portion(1,2). Together, 20S and 19S form the 26S proteasome, a protein complex responsible not only for protein degradation but also cell-cycle control, genetic regulation, DNA repair and immune response(1).
The proteasome is also involved in the regulation of memory(2-6): it is required to allow long-term memory formation(3,4) and reconsolidation(5), as well as mediate spine outgrowth(6), influencing synaptic plasticity. The UPS is therefore a highly investigated pathway in neurodegenerative disorders, such as Alzheimer’s disease (AD)(7-9). Other than memory loss and cognitive deficit, one of the many features of AD is chronic neuroinflammation, which can be observed in post-mortem samples and mouse models of AD(10). Yet, the link between proteasome activity, memory deficits and inflammation has not been well established to this date, which is exactly what this preprint aims to do.
Key findings
Genetically disrupting the 26S proteasome reduces proteasome activity and impairs animal learning and memory
The preprint authors evaluated proteasome activity in all three proteolytic subunits. All activities were decreased in Psmc1 KO mice when compared to the control group. During the Radial Arm Water Maze (RAWM) test (which evaluates memory), KO mice made more mistakes while trying to find the platform and spent more time searching for it. This result suggests that impairing 19S proteasome function causes learning and memory deficits in mice.
26S proteasome deficiency causes accumulation of inflammation-associated proteins
Using mass spectrometry, Psmc1 KO mice showed alterations in immune system-related functions. Given that result, the authors investigated the expression of proteins associated with inflammation, namely STAT1, TREM2 and NF-kB, as well as glial activation markers GFAP and Iba-1. Psmc1 KO mice showed increased protein levels of all targets, suggesting that this knockout mouse in fact has a higher inflammatory burden.
Inhibition of NF-κB improves learning and memory in Psmc1 KO animals
To investigate if NK-kB mediates the observed memory impairment, the authors injected the mice with PDTC, a NF-kB inhibitor, daily for three weeks. NF-kB inhibition was able to reverse the memory deficit caused by proteasome activity impairment after the 8th block of experiments.
Inhibition of NF-κB improves 26S proteasome deficiency-caused neuroinflammation
After injections with PDTC for three weeks, forebrain samples of Psmc1 KO treated mice were prepared and the protein content was analyzed by western blotting. The inhibition of NK-kB not only prevented the increase of inflammatory markers observed in Psmc1 KO but also reversed it, displaying an apparent general anti-inflammatory effect.
Graphical abstract:
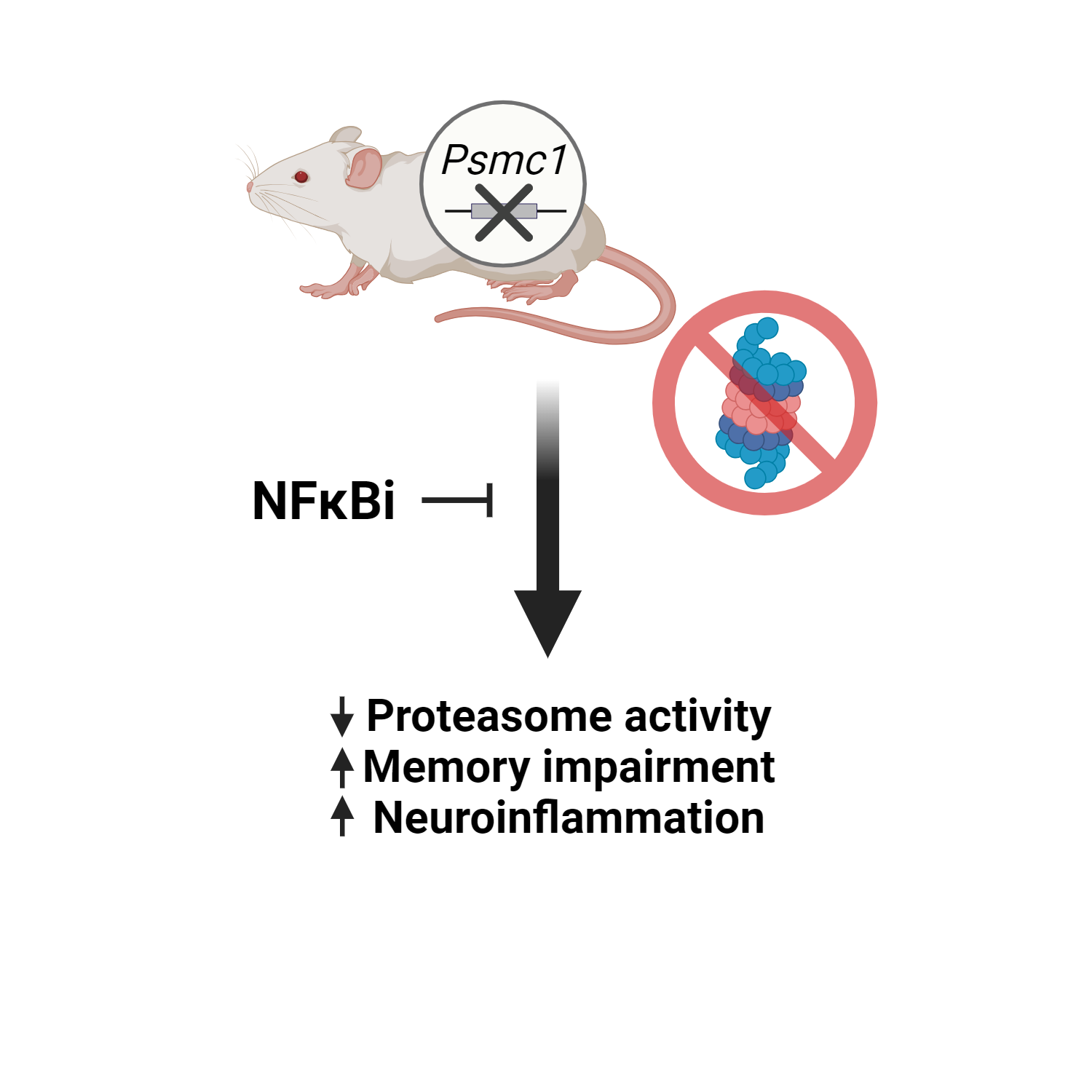
Figure 1: Graphical summary of the Huber et al. preprint showing the evaluated effects of Psmc1 KO in mice. Illustration made with Biorender.
Why I think this preprint is important
This preprint links the cognitive deficit caused by forebrain-specific Psmc1 KO with inflammatory mediators, suggesting that the UPS is responsible at least partially for regulating inflammation. This finding is not only relevant to inflammation and memory in the forebrain, but also to other cognitive related areas and cellular functions. In addition, this is to our knowledge the first research project that used Psmc1 knock out animals to study memory loss, a classic AD symptom, linking this specific proteasome gene to an AD-like phenotype.
Questions and suggestions
Q1: In the “Materials and Methods” section, it would be important to include the following details about the animals: the Ethics Committee protocol number, the sex of the animals, and their lineage and sub lineage (Enriquez et al., 2019). Additionally, it would be good to provide information about the temperature and general conditions of the animal facility. These details are crucial as they can significantly impact the results, particularly concerning the genetic background of both control and KO animals, which should be kept as similar as possible.
Q2: In the first set of experiments, the authors measured proteasome activity in the forebrain of KO mice. Given the fact that RAWM is a cognitive test that evaluates spatial learning and areas such as the hippocampus or entorhinal cortex are more associated with this type of learning, measuring the proteasome activity in those brain regions could be more interesting when making this association with the RAWM results. Considering that the authors knocked out Psmc1 only in the forebrain (which is related to working memory),
perhaps a Y-maze would be a more suitable test, since it evaluates working memory and others, becoming a more suitable cognitive test(11).
Q3: Still regarding the RAWM experiments, were the RAWM results shown in Figure 1 performed in parallel with the ones shown in Figure 4? If yes, it would be interesting to include the control group results in Figure 4 B and C to evaluate if the group performance on this test was different when compared to KO + PDTC. By doing this, the authors can discuss whether PDTC completely or partially reverts the memory impairment caused by Psmc1 KO.
Q4: In the “Synaptosome isolation” method description, there is a typographical error in the first sentence (“tperformed”).
Q5: In the description of the Mass spectrometric analysis of synaptic proteins, it is worth mentioning that this experiment is essentially a proteomic analysis in the “Material and methods” section.
Q6: Could the authors check that all graphs contain the unit of the measurement in the y-axis? Most of the figures do not display “% of control” or “relative to control”
Q7: As part of Figure 3, ELISA protocols to detect proinflammatory cytokines such as IL-1β or IFN-γ could be interesting to confirm the inflammatory state caused by Psmc1 KO since the synthesis and liberation of cytokines are downstream events that confirm the inflammatory state. Although TREM2 and STAT1 do suggest inflammation, additional data would help to support these results. TREM2, for example, is highly expressed in disease-associated microglia (DAM), but not necessarily a marker of inflammation(12). The same for Iba-1 and GFAP, which by themselves imply cell activation. Also, it appears that the antibody used in Figure 3D stains total NF-Kb. It would perhaps be more suitable to use p65 NF-kB, which stains the active form of NF-kB, or at least evaluate if this nuclear factor collocates with DAPI in an immunohistochemistry assay (if it is indeed in the nucleus), which could be another indication that NF-kB is in fact inducing inflammation.
Q8: Currently, the immunohistochemistry legends only inform the reader that the experiments were performed in the cortex. It should perhaps be made more explicit what brain region was sliced and stained. Considering the experiments were done in total cortices, it is probably more suitable to stain only forebrain samples.
Q9: In the PDTC injection experiments, shown in Figure 4, it would be beneficial to include “control” and “control + PDTC” groups to ensure that the inhibitor is reversing the effect of Psmc1 KO. PDTC could by itself provide a general improvement of memory and surely would decrease inflammatory markers evaluated in Figure 5, so the suggested additional groups can be included to strengthen the authors’ conclusions. In addition, although PDTC has been described as a selective NFkB inhibitor, it may have off-target effects that should be taken in consideration when discussing the results.
Q10: One additional suggestion is to repeat the western blotting experiments loading the samples used in Figures 3 and 4. By doing this, the authors will be able to compare more experimental groups, and this could provide additional data to enrich the discussion of whether PDTC reverts or attenuates the effects of Psmc1 KO in the pathways and inflammatory/reactivity markers evaluated in these experiments.
Q11: The authors could consider including full western blotting membranes as Supplementary Figures to ensure that unspecific binding of antibodies does not take place in their experiments. This transparency increases the reliability of the data and avoids further questions.
Q12: In the current manuscript, the authors frequently mention AD but none of the results displayed are directly related to AD models. To bring AD into the discussion, the authors could perform at least one experiment that links Psmc1 KO with AD. One relatively simple suggestion is to harvest astrocytes and microglia of wild type and KO animals in culture dishes and treat them with amyloid-β oligomers. The comparison of protein expression between these two groups would allow the authors to observe the effects of the KO in a widely used AD model. If this or similar experiments are not possible, it would perhaps be a good idea to rephrase the parts of the discussion that mention AD since memory loss is only one symptom of a much more complex disease.
Q13: Recently, Psmc1 KO was correlated to a rare neurological syndrome named Birk-Aharoni Syndrome (BKAS)(13). This autosomal recessive syndrome causes phenotypic alterations such as impaired general development, intellectual disability, spastic tetraplegia, hearing loss, male genital alterations and elevation of liver enzymes. Do the Psmc1 KO mice have any phenotype similarities to the ones described by Birk and Aharoni? If yes, the results displayed in this paper might have additional contributions that go beyond AD.
References:
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009; 10:104–115. doi: 10.1038/nrm2630
- Ribeiro FC, Cozachenco D, Heimfarth L, Fortuna JTS, de Freitas GB, de Sousa JM, Alves-Leon SV, Leite REP, Suemoto CK, Grinberg LT, De Felice FG, Lourenco MV, Ferreira ST. Synaptic proteasome is inhibited in Alzheimer’s disease models and associates with memory impairment in mice. Commun Biol. 2023 Nov 7;6(1):1127. doi: 10.1038/s42003-023-05511-9. PMID: 37935829; PMCID: PMC10630330.
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem. 2008 Apr 25;15(5):335-47. doi: 10.1101/lm.984508. PMID: 18441292; PMCID: PMC2364605.
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001 Dec;14(11):1820-6. doi: 10.1046/j.0953-816x.2001.01806.x. PMID: 11860477.
- Artinian J, McGauran AM, De Jaeger X, Mouledous L, Frances B, Roullet P. Protein degradation, as with protein synthesis, is required during not only long-term spatial memory consolidation but also reconsolidation. Eur J Neurosci. 2008 Jun;27(11):3009-19. doi: 10.1111/j.1460-9568.2008.06262.x. PMID: 18588539.
- Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, Zito K. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012 Jun 21;74(6):1023-30. doi: 10.1016/j.neuron.2012.04.031. PMID: 22726833; PMCID: PMC3500563.
- Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000 Jul;75(1):436-9. doi: 10.1046/j.1471-4159.2000.0750436.x. PMID: 10854289.
- Harris LD, Jasem S, Licchesi JDF. The Ubiquitin System in Alzheimer’s Disease. Adv Exp Med Biol. 2020;1233:195-221. doi: 10.1007/978-3-030-38266-7_8. PMID: 32274758
- Li Z, Jansen M, Pierre SR, Figueiredo-Pereira ME. Neurodegeneration: linking ubiquitin/proteasome pathway impairment with inflammation. Int J Biochem Cell Biol. 2003 May;35(5):547-52. doi: 10.1016/s1357-2725(02)00384-9. PMID: 12672447.
- Fakhoury M. Inflammation in Alzheimer’s Disease. Curr Alzheimer Res. 2020;17(11):959-961. doi: 10.2174/156720501711210101110513. PMID: 33509069.
- Kraeuter AK, Guest PC, Sarnyai Z. The Y-Maze for Assessment of Spatial Working and Reference Memory in Mice. Methods Mol Biol. 2019;1916:105-111. doi: 10.1007/978-1-4939-8994-2_10. PMID: 30535688.
- Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell. 2018 May 17;173(5):1073-1081. doi: 10.1016/j.cell.2018.05.003. PMID: 29775591.
- Aharoni S, Proskorovski-Ohayon R, Krishnan RK, Yogev Y, Wormser O, Hadar N, Bakhrat A, Alshafee I, Gombosh M, Agam N, Gradstein L, Shorer Z, Zarivach R, Eskin-Schwartz M, Abdu U, Birk OS. PSMC1 variant causes a novel neurological syndrome. Clin Genet. 2022 Oct; 102(4):324-332. doi: 10.1111/cge.14195. Epub 2022 Aug 3. PMID: 35861243; PMCID: PMC9541193.
doi: https://doi.org/10.1242/prelights.38193
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the animal behavior and cognition category:
Cannibalism as a mechanism to offset reproductive costs in three-spined sticklebacks
Tina Nguyen
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Also in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the neuroscience category:
Electrophysiological correlates of conscious experiences during sleep: Lucid dreams, sleep paralysis, out-of-body experiences, and false awakenings
uMontreal Neuro preLighters et al.
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
preLists in the animal behavior and cognition category:
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Also in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)