A novel mechanism of gland formation in zebrafish involving transdifferentiation of renal epithelial cells and live cell extrusion
Posted on: 17 July 2018
Preprint posted on 22 June 2018
Article now published in eLife at http://dx.doi.org/10.7554/elife.38911
A change of identity drives organogenesis: a rare event of transdifferentiation in zebrafish.
Selected by Giuliana ClementeCategories: developmental biology
Context and Background:
Can the differentiated state really be considered terminal and irreversible? This was the challenge that Helen Blau took up in the mid-80s when, inspired by the work of John Gurdon, she set up to unhinge the dogma of terminal differentiation according to which the gene expression profile of a specialised cell is irreversible.
Blau was the first researcher to talk about cell plasticity. She proposed that the differentiated state, albeit stable, is not fixed and that cells can potentially change identity by re-activating and/or silencing specific sets of genes in a dynamic fashion.
This new concept has two important implications. First, the differentiated state is actively controlled and a constant regulation holds it in place. Secondly, terminally differentiated cells can acquire a new fate, adapting to changes in the environmental conditions, in a process known as transdifferentiation (1,2).
From an experimental point of view, the idea of plasticity led to the reasoning that somatic cells can be forced to reverse back to a stem cell-like state or into a different cell lineage by playing around with the expression levels of transcription factors. For example, in 1987 Tapscott and colleagues reported that expression of MyoD in fibroblasts was enough to force cells directly into the muscle cell lineage (3). Almost 20 years later, Yamanaka was able to revert skin cells of adult mice into pluripotent stem cells (iPS) by using a cocktail of the 4 transcription factors Sox-2, Klf-4, Oct-3/4, c-Myc (4). This pioneering work, that won Yamanaka the Nobel Prize, revolutionised the field of regenerative medicine, paving the way to more personalised therapies, disease modelling and drug discovery.
Although it is possible to achieve transdifferentiation in vitro, examples of this process in an in vivo context are rare and they normally occur upon injury or in pathological settings such as cancer. Even more unusual are examples of transdifferentiation during development and the extent to which the process contributes to organogenesis is still somewhat elusive.
Key findings:
In this preprint, Naylor and Davidson describe a rare event of transdifferentiation involved in the formation of the endocrine gland Corpuscles of Stannius (CS) in zebrafish. They show that the CS originates from a subset of cells of the distal early (DE) renal tubular epithelium (Figure 1A). The process of CS formation can be divided into two steps: the initial transdifferentiation of the DE cells into CS cells and the subsequent extrusion of the CS cells from the renal epithelium. The change in cell fate is under the control of the Notch signalling pathway. Notch promotes the switch from DE to CS lineage by promoting the export of Hnf1b, master regulator of the renal fate, from the nucleus to the cytoplasm (Figure 1B). This is the first time that nuclear export and sequestration of a transcription factor are shown to drive transdifferentiation. This working model is supported by experimental data obtained by nicely combining genetic and chemical approaches. Specifically, the authors show that morpholino or CrispR-Cas9 for irx3b (downstream effector of Hnf1b) result in an increased number of stc1+ cells and decreased nuclear localisation of Hbn1b. This observation suggests that the Hbnf1-irx3b network negatively regulates transdifferentiation. Conversely, the researchers observed a reduction in the number of CS cells specified in mindbomb mutants, defective in Notch, as well as upon the treatment with the g–secretase inhibitor cpdE, a result that places Notch as positive regulator of the DE-CS transition. As a final proof, an epistasis experiment was performed which consisted in treating irx3b crispants with cpdE. The inhibition of Notch rescued the loss of irx3b, resulting in less CS cells and the retention of Hnb1f in the nucleus.
The second step of the process is the extrusion of the CS cells from the renal tubule. The extrusion, which is driven by acto-myosin dependent apical constriction of the CS cells as proved by Blebbistatin treatment, is later followed by cell proliferation and clonal expansion of the small group of CS cells.
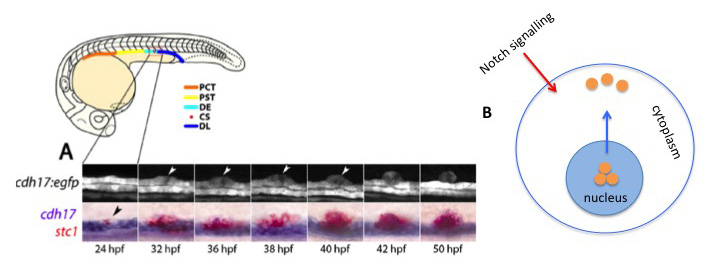
Figure 1: Origin of the Corpuscles of Stannius (CS) in zebrafish
A) The endocrine gland forms from the most posterior portion of the DE tubule by a process of transdifferentiation. At 24 hours post fertilisation (hpf), a small group of cells in the DE downregulate the expression of the renal marker cdh17 and at the same time start expressing stanniocalcin-1 (stc1). 50 hpf, the stc1-positive cells form a discrete structure above each pronephric tubule. B) The process of transdifferentiation is positively regulated by Notch signaling. Notch promotes the shuttling (blue arrow) of the transcription factor Hnf1b (orange) from the nucleus to the cytoplasm. Picture in (A) was adopted from figure 1 of the preprint.
Why I chose this paper:
We came a long way down the original idea that cells at their latest point of differentiation are unable to reverse to a progenitor state or change into another cell type. While cells can be forced experimentally to acquire a new identity, we now know that transdifferentiation occurs in nature under physiological conditions as well, for example during morphogenesis or tissue repair as a response to cell loss. Moreover, this phenomenon has been implicated in the onset of various forms of cancer but it can also be associated to other pathologies such as liver fibrosis and cirrhosis.
How transdifferentiation occurs at a molecular level is still a matter of intense research. Clearly, a better characterisation of the process will benefit our understanding of development, regeneration and diseases aetiology. Moreover, transdifferentiation harbours a great therapeutic potential and being able to properly manipulate cell identity is very appealing in regenerative medicine.
In this work the authors uncover a previously uncharacterised event of transdifferentiation in the developing zebrafish embryo. As such, the system represents a new model to study the physiological relevance of transdifferentiation in vivo and will allow to better address the molecular basis of cell plasticity.
Future directions and Questions to the authors:
The preprint leaves the readers with a lot of interesting open questions on how the DE-DC transition is regulated in space and time.
- At what time point during development does the Notch signalling hit the threshold to overcome the Hnf1b-irx3b regulation?
- How is the initial number of CS cells set? The Notch signalling could promote the transdifferentiation and at the same time prevent it from happening in the neighbouring cells by lateral inhibition.
- Is it Notch controlling the extrusion of the CS cells from the renal epithelium?
References:
- Blau, H.M., Pavleth, G.K., Hardeman, E.C. et al. (1985). Plasticity of the differentiated state. Science 230: 758–766.
- Blau, H.M. and Baltimore, D. (1991) Differentiation requires continuous regulation. Journal of Cell Biology 112: 781–783.
- R.L. Davis, et al.-Expression of a single transfected cDNA converts fibroblasts to myoblasts- Cell, 51 (1987), pp. 987-1000.
- Takahashi, K. and Yamanaka, S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676.
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)