Tumor-infiltrating CD27-IgD- regulatory B cells suppress cytotoxic CD8+T cell responses in renal cell carcinoma
Posted on: 12 August 2025 , updated on: 3 November 2025
Preprint posted on 12 July 2025
Categories: bioinformatics, immunology, molecular biology
Background
The function of B cells is highly dependent on the specific cancer type and its unique tumor microenvironment (TME). Typically, having a large number of B cells in a tumor is a good sign and leads to a higher chance of survival [1]. However, in renal cell carcinoma (RCC), the opposite seems to be true, with more B cells leading to worse outcomes.
Recent pan-cancer atlases have started to map out and investigate the multiple functions of tumor-infiltrating B cells (TIBs) by combining data from many different cancers [2] [3] [4]. However, these large-scale studies are often biased towards cancers with high B cell numbers, which can mask critical differences in tumors with low B-cell infiltration, like renal cell carcinoma (RCC). In this paper, Baig et al. use a novel single-cell method, combined with flow cytometry and high-plex imaging, to identify a previously unrecognized double negative 1 (DN1) B-cell subset (“double-negative” because these B cells lack the usual IgD and CD27 markers) that is enriched in RCC and is heavily associated with worse survival.
Key Highlights
Double Negative 1 (DN1) regulatory B cells (Breg) are enriched in RCC tumors
The authors used a unique clustering method of scRNA-seq data by first clustering B cells within each of four cancer types (breast, colon, lung, and RCC) before projecting them into a shared space for comparison. This method allowed them to identify B cell clusters in different cancers. Notably, they identified a new B cell population called double-negative 1 (DN1) B cells. Using a new integration model, known as the single-cell Variational Inference (scVI), the authors found that DN1 B cells are more common in RCC than other cancers. Specifically, DN1 B cells constitute 35.7% of all tumor-infiltrating B cells in RCC, while DN1 B cells constitute 1% of the other cancers.
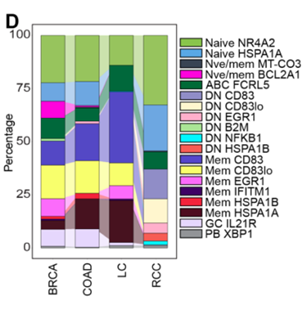
Figure 1D from the preprint – Percentage of different B cell subtypes in each of the four cancers studied (BRCA, COAD, LC, and RCC).
DN1 B cell enrichment is associated with worse survival
The researchers collected tumor samples from patients with RCC undergoing surgery (nephrectomy). These patients had not yet received treatment prior to surgery. They ran flow cytometry on these samples, which confirmed a significant increase in DN1 B cells in tumor tissue.
The researchers linked B cell enrichment to worse survival by using a validated Leibovich score to predict the risk of cancer recurrence after nephrectomy. They found that high-risk patients exhibit increased levels of DN1 B cells, DN3 B cells, and plasma cells.
Additionally, researchers used an adjusted multivariable Cox proportional hazards model to analyze a large public database (TCGA). From this model, they showed that three gene signatures for these regulatory B cells (CD19, IL10, TGFB1) were associated with poorer patient survival, as shown in Figure 5I.
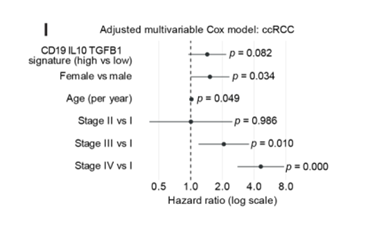
Figure 5I from the preprint – This figure is a forest plot showing that in a large database of RCC patients gathered from The Cancer Genome Atlas (TCGA), a high gene signature score for these regulatory B cells (CD19, IL10, TGFB1) was associated with worse survival.
DN1 Breg cells are found in tertiary lymphoid structures (TLSs) in RCC tumors
Researchers conducted high-plex spatial imaging of regions of interest (ROIs) to determine tertiary lymphoid structures (TLSs). They used cell segmentation and found that DN B cells are the most abundant B cell population within these structures. They found that the DN1 subset was the most frequent out of all DN B cells identified.
Naive B cells differentiate into DN1 Breg cells due to Toll-like receptor (TLR) signaling
The authors conducted a gene set enrichment analysis (GSEA) of the scRNA-seq data and found that Toll-like receptor (TLR) signaling pathways were upregulated. GSEA further revealed that the genes TLR7 and TLR9 specifically were enriched within the tumor’s DN1 B cell clusters. The authors then validated these findings using in vitro stimulation assays of TLR7 and TLR9 ligands. From these analyses, the authors were able to conclude TLR’s role in promoting B cell differentiation.
DN1 Breg cells promote reprogramming of CD8+ T cells into a T-regulatory (i.e. more tumor-friendly) phenotype
The authors conducted cell-cell interaction and found that B cells mostly engage with CD8+ T cells compared to other immune cell subsets. Spatial profiling further showed that Bregs are located close to CD8+ T cells. In vitro experiments revealed that tumor-derived DN B cells co-cultured with cytotoxic CD8+ T cells significantly reduced the T cells’ ability to produce the key anti-tumor cytokine IFN-γ.
Overall, Baig et al. used a variety of computational and experimental analysis to identify a new, enriched B cell population within RCC tumors. Further experimental analysis showed that these DN1 B cells are enriched in TLSs, and their abundance is associated with higher risk and poorer survival. They are developed from naïve B cells via TLR7/9-driven differentiation. Functionally, these DN1 Bregs engage nearby CD8+ T cells to suppress IFN-γ and bring these T cells toward a more tumor-permissive state.
Why this paper is important
One notable strength of this paper is the authors’ multi-modal approach to discover a new cell population (DN1 Bregs) responsible for immunosuppression in RCC. The authors first used a unique bioinformatic workflow to identify an immunosuppressive DN1 Breg population in RCC, then confirmed this finding’s significance using flow cytometry, spatial imaging, and in vitro functional assays.
I thought the scRNA-seq pipeline that the authors took was especially unique and inventive. Their innovative pipeline of clustering and integrating B cells within different cancer types and implementing a deep-learning model for projection allowed them to have more detailed cell clusters compared to the regular scRNA-seq pipeline and even led them to discover a new and important B cell cluster.
References
[1] Liu, H., Li, Z., Han, X., Li, Z., Zhao, Y., Liu, F., Zhu, Z., Lv, Y., Liu, Z., and Zhang, N. (2023). The prognostic impact of tumor-infiltrating B lymphocytes in patients with solid malignancies: A systematic review and meta-analysis. Critical Reviews in Oncology/Hematology 181, 103893. https://doi.org/10.1016/j.critrevonc.2022.103893.
[2] Fitzsimons, E., Qian, D., Enica, A., Thakkar, K., Augustine, M., Gamble, S., Reading, J.L., and Litchfield, K. (2024). A pan-cancer single-cell RNA-seq atlas of intratumoral B cells. Cancer Cell 42, 1784-1797.e4. https://doi.org/10.1016/j.ccell.2024.09.011.
[3] Yang, Y., Chen, X., Pan, J., Ning, H., Zhang, Y., Bo, Y., Ren, X., Li, J., Qin, S., Wang, D., et al. (2024). Pan-cancer single-cell dissection reveals phenotypically distinct B cell subtypes. Cell 187, 4790-4811.e22. https://doi.org/10.1016/j.cell.2024.06.038.
[4] Ma, J., Wu, Y., Ma, L., Yang, X., Zhang, T., Song, G., Li, T., Gao, K., Shen, X., Lin, J., et al. (2024). A blueprint for tumor-infiltrating B cells across human cancers. Science 384, eadj4857. https://doi.org/10.1126/science.adj4857.
doi: https://doi.org/10.1242/prelights.41204
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Computational design of pH-sensitive binders
Mohammed JALLOH
Longitudinal single cell RNA-sequencing reveals evolution of micro- and macro-states in chronic myeloid leukemia
Charis Qi
Also in the immunology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Scalable transcription factor mapping uncovers the regulatory dynamics of natural and synthetic transcription factors in human T cell states
Inês Caiado
Also in the molecular biology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the immunology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)
1 week
Tommaso
This PreLights highlight on tumor-infiltrating CD27^- IgD^- regulatory B cells in renal cell carcinoma makes an important contribution to our understanding of how specific B-cell subsets can exert immunosuppressive effects on cytotoxic CD8⁺ T cells within solid tumors. The identification of a pro-tumorigenic Breg population that locates to tertiary lymphoid structures and impairs effector T-cell function aligns with emerging evidence across cancer types that intratumoral B cells can adopt regulatory phenotypes with significant clinical implications.
In lung cancer, previous and complementary observations have also been reported (missed in your and Mauri et al referencies). For example, metabolically active and highly polyfunctional VISTA⁺ regulatory B cells were shown to be enriched in the tumor microenvironment of early-stage NSCLC and correlate with tumor recurrence, providing independent evidence that regulatory B subsets can shape anti-tumor immunity in human carcinoma. Although the regulatory mechanisms and marker profiles partially differ (e.g., VISTA expression and metabolic adaptation versus CD27^- IgD^- phenotype), both studies highlight the suppressive potential of specialized B-cell subsets on cytotoxic T-cell responses. Additionally, deep phenotyping work in NSCLC patients has demonstrated that defects in regulatory B-cell repertoires, including subsets characterized by CD27dim and IL-10 expression, are associated with clinical outcomes such as immune-related toxicity following PD-1/PD-L1 checkpoint blockade, further underscoring the clinical relevance of Breg heterogeneity across tumor contexts.