Growth factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development
Posted on: 19 January 2020
Preprint posted on 27 December 2019
Article now published in eLife at http://dx.doi.org/10.7554/eLife.56079
FGF4 signalling in the blastocyst: A simple system for an important binary cell fate decision.
Selected by Pierre Osteil, Irepan Salvador-MartinezCategories: developmental biology
Background:
During the development of the vertebrate embryo, its cells must divide and specialise into different cell types. The first specialisation occurs when cells take a binary decision: to form the embryo body or its supporting tissues such as the placenta or the yolk sac. Despite decades of work, some mechanisms involved in this early transition remain poorly understood. It is unclear how the embryo controls the proportions of cells when proliferating. Especially given the observation that after modifying the ratio of progenitor cells, the proportions revert to normal after some time.
As a paradigm of this early cell fate decision in mammals, the team chose to address the preimplantation mouse embryo. The preimplantation embryo first multiplies the number of cells to form a mass (called morula) until the first specialisation: its internal cells become the inner cell mass (ICM) while the outer cells become the trophectoderm (TE; the progenitor cells of the placenta). The ICM then further specialises into the epiblast (Epi) and the primitive endoderm (PrE). The epiblast is comprised of the progenitors of all cells in the adult body, while the PrE cells contribute to both extraembryonic structures (the placenta and yolk sac) and the definitive endoderm (progenitors of the gastrointestinal tract).
This study was built upon two observations made during the development of mammalian embryos: the development of the embryo is not affected by 1) removing cells at early stages for preimplantation diagnosis in human and 2) adding cells in the mouse embryo during the generation of chimeras (by injecting embryonic stem cells – ESC) These findings led the team to the hypothesis that an internal signalling pathway could be controlling the cell fate and lineage sizes in the blastocyst, reflecting on its adaptability during early development.
The team previously showed that the three lineages forming the blastocyst (TE, Epiblast and PrE) are comprised of a consistent proportion of lineage-derived cells, suggesting a robust regulation of the cell population (Saiz et al. 2016). Previous studies have proposed that a direct mutual inhibition between NANOG (expressed in the ICM) and GATA6 (expressed in the PrE), could be responsible for the second stage of cell specialisation (Bessonnard et al. 2014; Huang et al. 2007; Nissen et al. 2017; Schröter et al. 2015; Tosenberger et al. 2017, 2019). However, the expression of Gata6 is supported by the MAPK pathway which is activated by FGF4 but the epiblast fate is obtained from a low FGF environment (Chazaud et al. 2006). Taken together, these studies led the authors to the hypothesis that FGF4 alone, rather than a direct inhibitory circuit between NANOG and GATA6, could be responsible for the rapid cell fate switch between the two cell types during the second cell specialisation.
To test this hypothesis, the authors designed an in silico model for cell proportions and differentiation, generated chimeras to build up the cell number in the embryo and laser dissection to reduce it and finally demonstrated that FGF4-controlled local gradients are solely responsible for the progenitors to undergo specialisation into PrE or Epi.
The results:
Chimeras were formed by aggregating WT GFP+ embryonic stem cells (ESC) into label-free Gata6-/- embryos. As the PrE requires the expression of Gata6, the Gata6-/- host cells cannot form the PrE. However, when WT ESCs are injected into the embryo, they can specialise into PrE and rescue its development. Interestingly, the authors noticed the fate of WT cells depended on the final ratio of WT to Gata6-/- cells in the embryo. Indeed, if the injected WT cells accounted for less than or equal to 40% of the total cell number, WT cells only formed PrE. In contrast, if the WT cells accounted for more than 40% of the total number of cells, they contributed to both the epiblast and PrE. This data suggests a non-cell autonomous mechanism for cell fate decision, or, in simpler words, that cell fate is decided based on the cell neighbourhood activity.
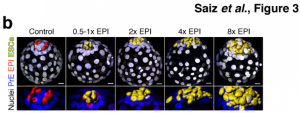
To gain a better understanding of the robustness of the ICM fate decision in a WT scenario, the team aggregated increasing amounts of fluorescently-labelled WT ESCs with unlabelled WT host embryos, to evaluate the effects of the varying proportion of ESC- to host-derived EPI cells (from 0.5x to 8x the host EPI cells). The ESC injection was performed at the 8-cell morula stage, before the first specialisation of the host blastomeres into ICM and TE. Interestingly, the number of cells injected did not affect the number of host-derived ICM cells indicating a cell-autonomous differentiation. However, it did impact the cell fate from the second specialisation event of the ICM; the more ESCs added, the more the host embryonic cells shifted away from the epiblast towards a PrE fate, confirming that the second specialisation was not cell-autonomous and instead based on the ratio of cells in the blastocyst.

To gain a more precise spatiotemporal control of lineage size manipulation, Saiz and colleagues optimised laser cell ablation to eliminate any desired cell in the blastocyst. Using this method, they observed a striking recovery; even after ablating either all Epi or PrE cells, all embryos contained cells of both lineages after 16-20 hours. However, this ability was lost at later developmental stages demonstrating a decline in the adaptability of the embryo as it develops. It has been demonstrated that FGF4 is involved in ICM specialisation into PrE. Saiz and collaborators hypothesised that FGF4 is essential in the growth factor-mediated feedback they proposed in their minimal mathematical model. Supporting this hypothesis, Fgf4-/- embryos do not form PrE and can be rescued by injecting WT ESCs with normal Fgf4 expression. Interestingly, the size of the resulting PrE was directly proportional to the number of WT ESCs injected.
In summary, the authors propose that the ratio of Epi versus PrE cells – rather than the absolute cell number – is an important determinant for cell fate, and that the expression of FGF4 is enough to regulate the Epi versus PrE cell fate decision.
Why did I choose this article?
I chose this article for the quality of the experiments performed and the amount of new insights gleaned by the team. It shows how flexible (hence impressive) the mammalian embryo is but also its limitations, giving the reader a clearer idea of its adaptability spectrum: one must add or remove a lot of cells to prevent the embryo to further develop. Furthermore, the team showed local concentrations of FGF4 ligand alone provide a fast and simple cue for cell fate changes. This is further supported by a simple mathematical model established by the authors. I enjoyed the simplicity of this model to explain the binary cell fate decision.
References:
Bessonnard, Sylvain, Laurane De Mot, Didier Gonze, Manon Barriol, Cynthia Dennis, Albert Goldbeter, Geneviève Dupont, and Claire Chazaud. 2014. ‘Gata6, Nanog and Erk Signaling Control Cell Fate in the Inner Cell Mass through a Tristable Regulatory Network’. Development (Cambridge) 141(19):3637–48. https://dev.biologists.org/content/141/19/3637.long
Chazaud, Claire, Yojiro Yamanaka, Tony Pawson, and Janet Rossant. 2006. ‘Early Lineage Segregation between Epiblast and Primitive Endoderm in Mouse Blastocysts through the Grb2-MAPK Pathway’. Developmental Cell 10:615–624. https://www.sciencedirect.com/science/article/pii/S1534580706001250?via%3Dihub
Huang, Sui, Yan Ping Guo, Gillian May, and Tariq Enver. 2007. ‘Bifurcation Dynamics in Lineage-Commitment in Bipotent Progenitor Cells’. Developmental Biology 305(2):695–713. https://www.sciencedirect.com/science/article/pii/S0012160607001674?via%3Dihub
Menchero, Sergio, Isabel Rollan, Antonio Lopez-Izquierdo, Maria Jose Andreu, Julio Sainz De Aja, Minjung Kang, Javier Adan, Rui Benedito, Teresa Rayon, Anna Katerina Hadjantonakis, and Miguel Manzanares. 2019. ‘Transitions in Cell Potency during Early Mouse Development Are Driven by Notch’. ELife 8:1–29. https://elifesciences.org/articles/42930
Nissen, Silas Boye, Marta Perera, Javier Martin Gonzalez, Sophie M. Morgani, Mogens H. Jensen, Kim Sneppen, Joshua M. Brickman, and Ala Trusina. 2017. ‘Four Simple Rules That Are Sufficient to Generate the Mammalian Blastocyst’. PLoS Biology 15(7):1–30. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.2000737
Rayon, Teresa, Sergio Menchero, Andres Nieto, Panagiotis Xenopoulos, Miguel Crespo, Katie Cockburn, Susana Cañon, Hiroshi Sasaki, Anna Katerina Hadjantonakis, Jose Luis de la Pompa, Janet Rossant, and Miguel Manzanares. 2014. ‘Notch and Hippo Converge on Cdx2 to Specify the Trophectoderm Lineage in the Mouse Blastocyst’. Developmental Cell 30(4):410–22. https://www.sciencedirect.com/science/article/pii/S1534580714004080?via%3Dihub
Saiz, Néstor, Kiah M. Williams, Venkatraman E. Seshan, and Anna Katerina Hadjantonakis. 2016. ‘Asynchronous Fate Decisions by Single Cells Collectively Ensure Consistent Lineage Composition in the Mouse Blastocyst’. Nature Communications 7. https://www.nature.com/articles/ncomms13463
Schröter, Christian, Pau Rué, Jonathan Peter Mackenzie, and Alfonso Martinez Arias. 2015. ‘FGF/MAPK Signaling Sets the Switching Threshold of a Bistable Circuit Controlling Cell Fate Decisions in Embryonic Stem Cells’. Development (Cambridge) 142(24):4205–16. https://dev.biologists.org/content/142/24/4205
Tosenberger, Alen, Didier Gonze, Sylvain Bessonnard, Michel Cohen-Tannoudji, Claire Chazaud, and Geneviève Dupont. 2017. ‘A Multiscale Model of Early Cell Lineage Specification Including Cell Division’. Npj Systems Biology and Applications 3(1):1–10. http://dx.doi.org/10.1038/s41540-017-0017-0
Tosenberger, Alen, Didier Gonze, Claire Chazaud, and Geneviève Dupont. 2019. ‘Computational Models for the Dynamics of Early Mouse Embryogenesis’. International Journal of Developmental Biology 63(3-4–5):131–42. http://www.ijdb.ehu.es/web/paper/180418gd/computational-models-for-the-dynamics-of-early-mouse-embryogenesis
doi: https://doi.org/10.1242/prelights.16272
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)