A patterned human heart tube organoid model generated by pluripotent stem cell self-assembly
Posted on: 13 February 2023 , updated on: 14 February 2023
Preprint posted on 20 December 2022
A patterned human heart tube organoid model generated by pluripotent stem cell self-assembly
Selected by Silvia BeccaCategories: bioengineering, developmental biology
Background:
Recent improvements in human organoid technology have gained substantial interest. “Organoids” represent a unique possibility to model human tissues and even organs starting from stem cells, particularly human induced pluripotent stem cells (hiPSCs). Some organoids recapitulate key physiological and metabolic functions. This makes them potentially suitable for a variety of applications ranging from developmental and disease modelling studies to drug screening and even cell therapy. Organoids also promise to overcome the ethical and biological limitations related to the use of animal models in many research areas.
Among other applications, organoids are an attractive model to study congenital heart defects. Due to the underlying complexity, this goal would ideally require reproducing the variety of cell types that are specified between the 4th to 6th week of human gestation, while also recapitulating various layers of organ architecture (Lewis-Israeli et al., 2022). Existing human heart organoid models have achieved the self-assembly of certain cell lineages derived from the first and second heart field (FHF and SHF) into chamber-like cave structures that can spontaneously contract (Drakhlis et al., 2021; Hofbauer et al., 2021b; Lewis-Israeli et al., 2022) . Despite the remarkable level of complexity achieved, these models do not yet replicate some key developmental milestones including heart tube polarization, looping, and vascularization (Hofbauer et al., 2021a).
The Aguirre laboratory previously published a cardiac organoid model comprising of several key cardiac cell types from both the FHF and SHF (approximately 59% cardiomyocytes, 16% epicardial cells, 14% endocardial cells, 12% cardiac fibroblasts, and 1.6% endothelial cells) (Lewis-Israeli et al., 2022). Nevertheless, they recognised that this model still lacked important cell types, like valve and conductance cells, and did not present right-left and anterior-posterior patterning. In this preprint the same group made important steps towards tackling these issues.
Major findings:
The goal of this study was to induce organoid maturation in order to more closely mimic the human heart at approximately gestational day 45 (GD45). To achieve this, Volmert and colleagues tested various culture conditions to better recapitulate the hormonal and metabolic changes that occur during in vivo heart development.
The authors gradually increased the complexity of the maturation medium, which was applied to cardiac organoids from day 20 to day 30 of their differentiation. Eventually, they identified a cocktail including fatty acids (FA) and L-carnitine provided in a low glucose medium that could recapitulate the physiological switch of the developing heart from glucose-based to FA-based metabolism. This was complemented by the temporally-controlled addition of two hormones with established roles in foetal heart growth and development: triiodothyronine (T3) and insulin growth factor 1 (IGF-1). All of these factors had been previously reported to improve ventricular cardiomyocyte maturation protocols (Karbassi et al., 2020), but both their application to cardiac organoids and the specific combination reported here is novel. The resulting protocol led to important effects on metabolic maturation, such as an increase in mitochondrial content and expression of genes involved in FA metabolism and oxidative phosphorylation (i.e. PPARA, UQCRB, PPARGC1, BNRF1).
Single cell RNA sequencing (sc-RNA-seq) experiments indicated the emergence of various cardiac cell types, particularly cardiomyocytes (CM) and stromal cells, but also proepicardial, valve, and conductance cells among other rarer cell types. Most notably, matured organoids showed the emergence of approximately 3 per 1000 epicardial cells, which while rare may have important functions, as discussed below. It is nevertheless worth noting that several key cardiac cell types seem to be still missing (i.e. cardiac neural crest cells, understandably given their ectodermal origin (George et al., 2020)) or greatly underrepresented (i.e. endothelial cells, which as noted by the authors are somehow reduced after maturation, which contrasts with their high prevalence and key developmental role in early cardiac development in vivo (Haack and Abdelilah-Seyfried, 2016)). Interestingly, the authors distinguished atrial and ventricular cardiomyocytes chiefly through the differential expression of sarcomeric protein isoforms (i.e. MYL7 vs MYL2/3 and MYH6 vs MYH7). Though, several such switches are most often regarded as indicators of developmental progression from immature to mature ventricular CMs (Karbassi et al., 2020), and alternative markers such as NR2F2 and IRX1/4 have been reported for atrial vs ventricular identity, respectively (Churko et al., 2018; Nelson et al., 2016; Schmidt et al., n.d.). Of note, the authors did not comment on whether the enhanced maturation protocol affects the specification of left versus right ventricular CMs, which arise from different embryonic progenitors in the FHF and SHF, respectively.
Having established their cardiac organoid model, the authors investigated its potential to replicate key changes in gene expression and electrophysiology during gestation. By comparing pseudo-bulk RNA seq with data from the Human Cell Atlas project (Asp et al., 2019), the authors proposed that their matured organoids are similar to the GD45 heart. For instance, there is comparable expression of certain epicardial cell and ventricular CM markers. It would be exciting to get a better understanding of the improvements driven by maturation by also comparing data at the single cell level. From the perspective of electrophysiology, the authors reported promising findings such as increased expression of mature cardiomyocytes ion channels and some T-tubule-like structures, even though their number is still low and it is unclear whether they are associated to RYR2 clustering (Parikh et al., 2017).
Remarkably, the authors reported having obtained, for the first time, some degree of anterior-posterior (A-P) patterning in the cardiac organoids. This is evident from immunofluorescence stainings that identified “atrial” and “ventricular” poles (Fig. 1). Moreover, they proposed that the atrial pole could be induced by an endogenous gradient of retinoic acid (RA). RA is known to be produced by (pro)epicardial cells in vivo, and it is a major morphogen involved in heart tube patterning (Duester, 2008). Accordingly, the atrial pole was found to contain a cluster of cells co-expressing the TBX18 (pro)epicardial marker and ALDH1A2, an enzyme responsible for converting retinol into retinoic acid. The presence of endogenous RA was further supported by Raman spectroscopy data. Matured organoids also exhibited a more elongated shape which reminisces the developing heart tube.
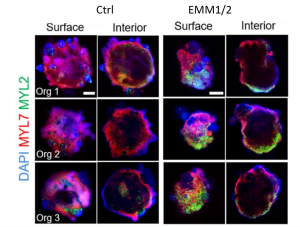
Fig 1. Developmental induction promotes the formation of well-developed atrial and ventricular chambers by self-organization. Representative surface and interior immunofluorescence images of individual day 30 organoids in control and EMM2/1 conditions. Three organoids are displayed for each condition (n=9-13 organoids per condition). DAPI (blue), MYL7 (red), MYL2 (green). Scale bars = 400 μm (Volmert et al., preprint).
Taken together with other results described in the preprint, the authors concluded having partially recapitulated the changes that occur during the first 4 to 6 weeks of development. They thus suggest that metabolically matured cardiac organoids could be a scalable model suitable for pharmacological testing applications.
What we liked about this preprint:
Heart tube organoids are an emerging and promising tool for several applications, but despite huge progress in the field we are still a long way for mimicking the more advanced morphogenetic changes involved in heart development: the A-P patterning reported in this preprint represents an important milestone towards this goal.
The RA gradient produced by the proepicardial pole, together with the WNT gradient, is a fundamental cue in in vivo heart development and contributes to the early specification of A-P patterning which drives the specification of atrial chambers versus ventricular ones. The most exciting outcome of this paper is that metabolic and hormonal maturation of organoids appears to be sufficient to support the differentiation and clustering of TBX18-ALDH1A2 at one pole of developing organoids, which may in turn drive an endogenous gradient able to break radial symmetry and drive A-P patterning of atrial and ventricular poles.
Questions to the authors:
- Early IGF-1 treatment appears to have a major impact on later morphogenetic events: any ideas on the mechanism involved?
- Do the atrial and ventricular cells derive from SHF and FHF progenitors, respectively, as during normal development? Alternatively, is the MYL7 to MYL2 gradient explained chiefly by selective maturation of FHF cells?
- Could the causal role of RA gradient in A-P patterning be established, perhaps through genetic or pharmacological inhibition?
- What is a key example of an application that would showcase the benefits of this model over earlier iterations?
- How reproducible is the protocol across different batches of differentiations and, most importantly, different iPSC lines? What are, if any, the reagents that require batch testing/dosage optimization?
Bibliography
Asp, M., Giacomello, S., Larsson, L., Wu, C., Fürth, D., Qian, X., Wärdell, E., Custodio, J., Reimegård, J., Salmén, F., Österholm, C., Ståhl, P.L., Sundström, E., Åkesson, E., Bergmann, O., Bienko, M., Månsson-Broberg, A., Nilsson, M., Sylvén, C., Lundeberg, J., 2019. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 179, 1647-1660.e19. https://doi.org/10.1016/j.cell.2019.11.025
Churko, J.M., Garg, P., Treutlein, B., Venkatasubramanian, M., Wu, H., Lee, J., Wessells, Q.N., Chen, S.-Y., Chen, W.-Y., Chetal, K., Mantalas, G., Neff, N., Jabart, E., Sharma, A., Nolan, G.P., Salomonis, N., Wu, J.C., 2018. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat. Commun. 9, 4906. https://doi.org/10.1038/s41467-018-07333-4
Drakhlis, L., Biswanath, S., Farr, C.-M., Lupanow, V., Teske, J., Ritzenhoff, K., Franke, A., Manstein, F., Bolesani, E., Kempf, H., Liebscher, S., Schenke-Layland, K., Hegermann, J., Nolte, L., Meyer, H., de la Roche, J., Thiemann, S., Wahl-Schott, C., Martin, U., Zweigerdt, R., 2021. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 39, 737–746. https://doi.org/10.1038/s41587-021-00815-9
Duester, G., 2008. Retinoic Acid Synthesis and Signaling during Early Organogenesis. Cell 134, 921–931. https://doi.org/10.1016/j.cell.2008.09.002
George, R.M., Maldonado-Velez, G., Firulli, A.B., 2020. The heart of the neural crest: cardiac neural crest cells in development and regeneration. Development 147, dev188706. https://doi.org/10.1242/dev.188706
Haack, T., Abdelilah-Seyfried, S., 2016. The force within: endocardial development, mechanotransduction and signalling during cardiac morphogenesis. Development 143, 373–386. https://doi.org/10.1242/dev.131425
Hofbauer, P., Jahnel, S.M., Mendjan, S., 2021a. In vitro models of the human heart. Development 148, dev199672. https://doi.org/10.1242/dev.199672
Hofbauer, P., Jahnel, S.M., Papai, N., Giesshammer, M., Deyett, A., Schmidt, C., Penc, M., Tavernini, K., Grdseloff, N., Meledeth, C., Ginistrelli, L.C., Ctortecka, C., Šalic, Š., Novatchkova, M., Mendjan, S., 2021b. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 184, 3299-3317.e22. https://doi.org/10.1016/j.cell.2021.04.034
Karbassi, E., Fenix, A., Marchiano, S., Muraoka, N., Nakamura, K., Yang, X., Murry, C.E., 2020. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 17, 341–359. https://doi.org/10.1038/s41569-019-0331-x
Lewis-Israeli, Y.R., Volmert, B.D., Gabalski, M.A., Huang, A.R., Aguirre, A., 2022. Generating Self-assembling Human Heart Organoids Derived from Pluripotent Stem Cells.
Nelson, D.O., Lalit, P.A., Biermann, M., Markandeya, Y.S., Capes, D.L., Addesso, L., Patel, G., Han, T., John, M.C., Powers, P.A., Downs, K.M., Kamp, T.J., Lyons, G.E., 2016. Irx4 Marks a Multipotent, Ventricular-Specific Progenitor Cell. Stem Cells 34, 2875–2888. https://doi.org/10.1002/stem.2486
Parikh, S.S., Blackwell, D.J., Gomez-Hurtado, N., Frisk, M., Wang, L., Kim, K., Dahl, C.P., Fiane, A., Tønnessen, T., Kryshtal, D.O., Louch, W.E., Knollmann, B.C., 2017. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell–Derived Cardiomyocytes. Circ. Res. 121, 1323–1330. https://doi.org/10.1161/CIRCRESAHA.117.311920
Schmidt, C., Deyett, A., Ilmer, T., Caballero, A.T., Haendeler, S., Netzer, M.A., Ginistrelli, L.C., Cirigliano, M., Mancheno, E.J., Tavernini, K., Hering, S., Hofbauer, P., Mendjan, S., n.d. Multi-chamber cardioids unravel human heart development and cardiac defects.
doi: https://doi.org/10.1242/prelights.33649
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioengineering category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Matrix viscoelasticity regulates dendritic cell migration and immune priming
Roberto Amadio
Also in the developmental biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the bioengineering category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (1 votes)
(1 votes)