Dynamic Aha1 Co-Chaperone Binding to Human Hsp90
Posted on: 19 February 2019
Preprint posted on 14 February 2019
Article now published in Protein Science at http://dx.doi.org/10.1002/pro.3678
Categories: biochemistry, biophysics
Background
Seemingly disparate diseases such as type II diabetes, cystic fibrosis, ALS, and Alzheimer’s share a common molecular hallmark, the misfolding of proteins1. However, cells have armed themselves with an evolutionarily conserved defense mechanism to combat protein misfolding: a class of proteins known as molecular chaperones that can help nascent proteins fold, refold incorrectly folded proteins, or recognize misfolded proteins and target them for degradation2. A key molecular chaperone is the 90 kDa heat shock protein (Hsp90), which is present in all eukaryotes and essential for cell viability3. A recent preLight on the molecular chaperone activity of Hsp90 can be found here.
Hsp90 is an ATP-dependent chaperone and therefore requires ATP hydrolysis for function3. In order to efficiently utilize the energy liberated from ATP hydrolysis for its molecular chaperone activity, the ATPase activity of Hsp90 is tightly regulated. The structural plasticity of Hsp90 provides such regulation: in its unbound form, the Hsp90 dimer populates an open V-shaped, with either ATP binding alone or the combined binding4 of co-chaperone and ATP promoting a transition to a closed conformation3. Furthermore, the Hsp90 dimer dynamically populates an equilibrium between multiple states (open, closed, intermediate), with co-chaperones and substrates shifting the equilibrium5. Over 20 Hsp90-interacting co-chaperones have been identified and these co-chaperones regulate the activity and substrate binding of Hsp903,5. Of these co-chaperones, however, only Aha1 is capable of stimulating Hsp90’s ATPase activity, signifying its crucial role as a regulator of Hsp90 activity.
Here, in this preprint6, the authors utilized nuclear magnetic resonance (NMR) spectroscopy to interrogate the interaction between human Hsp90 (HSP90β) and its co-chaperone Aha1. NMR spectroscopy is exquisitely sensitive to biomolecular interactions and molecular motions (see preLight here), rendering it a highly effective tool for the study of chaperones and their dynamic, plastic nature7. Due to the conformational gymnastics of Hsp90, many previous structural studies employed truncated constructs to simplify data collection and analysis. However, the preprint authors here make use of the full-length protein, thereby providing a more wholistic view.
Results
Previously, a crystal structure had been solved8 of a complex between the N-terminal region of Aha1 and the middle region of Hsp90. Structural data for the interaction between the two full-length proteins remained elusive and thus, since ATP is hydrolyzed in the N-terminal domain of Hsp90, it was unclear how Aha1 stimulated the ATPase activity of Hsp90. Here, the authors6 utilized the full-length proteins and NMR spectroscopy to gain a detailed and complete picture of this important interaction.
Because of the large size of the Hsp90-Aha1 complex (ca. 220 kDa), traditional NMR methods would be intractable due to very rapid NMR signal decay that hinders the study of molecules larger than ca. 50-60 kDa. Instead, the authors made use of a specific type of NMR spectroscopy called methyl-TROSY, which allows the study of biomolecules in excess of 1 MDa9. Methyl-TROSY requires 2H-labeled proteins that have selectively 1H, 13C-labeled methyl groups, which enables preservation of the NMR signals due to the favorable spectroscopic properties of methyl groups. Thus, only the methyl-bearing residues that have been isotopically labeled can yield NMR signals, lowering the number of probes for structural insight but also reducing spectral overlap. The authors here use isoleucine-labeled Hsp90, similar to prior studies of human Hsp90 by methyl-TROSY4,10,11,12.
With methyl-TROSY, the authors6 identified the Aha1 binding site on Hsp90, including widespread structural changes to the N-terminal domain of Hsp90, the site where ATP binds and gets hydrolyzed. When studying a biomolecular interaction by NMR, changes to NMR signals upon binding of a partner can manifest either from the direct binding event or from allosteric structural changes. It is well known that Hsp90 undergoes widespread structural changes upon binding to co-chaperones3,5, with allosteric rearrangements facilitating an “open” to “closed” transition. To disentangle the changes due to binding versus those from allostery, the authors made use of a truncated Hsp90 construct (Hsp90NM) lacking the C-terminal domain, which exists solely as a monomer. This allowed the authors to identify the outer area of the N-terminal domain (distant from the dimerization site) as an Aha1 binding site6.
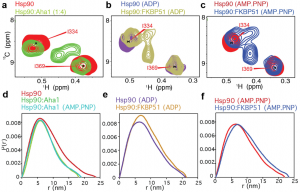
Figure 3 from the preprint. Shown in (a–c) are zoomed-in nuclear magnetic resonance (NMR) spectra (2D 1H-13C HMQC) of methyl-labeled Hsp90 (Ile-δ1 methyl groups are 1H, 13C labeled) in the (a) absence of nucleotide (red) bound to the co-chaperone Aha1 (green), which is NMR-invisible; (b) presence of ADP (purple) bound to the co-chaperone FKBP51 (gold), which promotes an open/extended conformation of Hsp90; or (c) presence of a non-hydrolyzable ATP analogue (AMP-PNP) (red) bound to FKBP51 (blue). (d–f) Small-angle X-ray scattering (SAXS) pair-distance distribution functions, P(r), for (d) Hsp90 (red), Hsp90-Aha1 (green), or Hsp90-Aha1 in the presence of AMP-PNP (cyan); (e) Hsp90 in the presence of ADP (gold) or Hsp90-FKBP51 in the presence of ADP (purple); and (f) Hsp90 in the presence of AMP-PNP (red) and FKBP51 (blue). This figure is reproduced here under a CC-BY-NC-ND 4.0 International license.
But how does Aha1 binding facilitate increased ATPase activity in Hsp90? An elegant study using yeast Hsp9013 reported both symmetric (2:2 Aha1:Hsp90) and asymmetric (1:2 Aha1:Hsp90) interactions involving Aha1 and Hsp90. Likewise, the stoichiometry of the interaction between co-chaperone p23 and Hsp90 was reported to be either a 1:2 ratio14 or 2:2 ratio4 of p23:Hsp90. NMR spectra are highly sensitive to symmetry, and the NMR data in this preprint are consistent with a symmetric interaction between two Aha1 molecules and the Hsp90 dimer6. A lowly populated asymmetric interaction cannot be ruled out, but the major state appears symmetric based on the single set of NMR signals that are present (e.g. Fig. 1b in the preprint). Finally, it is known that Hsp90 must transition to a closed conformation, placing the N-terminal domains in contact, for ATPase activity to be stimulated. The authors’ NMR data suggest that regions in the N-terminal domain undergo structural changes near the dimerization site, but their small-angle X-ray scattering data indicate that Hsp90 remains in a partially open/partially closed state6. Thus, the fully closed state is not formed and so it remains unclear if the N-terminal domains are in contact. Instead the data suggest that Aha1 binding leads to formation of an intermediate state, perhaps with transient contacts between N-terminal domains.
This study paves the way for future work investigating the inter-molecular contacts of the N-terminal domains of Hsp90 while bound to Aha1, as well as the mechanism of ternary complex formation involving Hsp90, Aha1, and other co-chaperones. Structural characterization of the Aha1-bound, dimer-like conformation of the Hsp90 N-terminal domain will prove insightful in understanding the mechanism of Aha1-induced stimulation of ATPase activity.
Why I chose this preprint
This preprint makes use of NMR spectroscopy to provide detailed insight into the mechanism of the co-chaperone Aha1 and its interaction with Hsp90. The first crystal structure of the truncated Hsp90-Aha1 complex appeared8 already in 2004, but difficulties in obtaining high-resolution data on the full-length complex prevented a wholistic view. Building upon and making use of previously solved crystal structures and prior biochemical and genetic assays, the new NMR data enable a powerful synthesis of data to aid in understanding the regulation of Hsp90 by Aha1.
References
- Chiti F, Dobon CM. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86: 27-68.
- Balchin D, Hayer-Hartl M, Hartl FU. (2016) In vivo aspects of protein folding and quality control. Science 353: aac4354.
- Schopf FH, Biebl MM, Buchner J. (2017) The HSP90 chaperone machinery. Nat. Rev. Mol. Cell. Biol. 18: 345-360.
- Karagoz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rudiger SG. (2011) N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc. Natl. Acad. Sci. U S A 108: 580-585.
- Sahasrabudhe P, Rohrberg J, Biebl MM, Rutz DA, Buchner J. (2017) The plasticity of the Hsp90 co-chaperone system. Mol. Cell 67: 947-951.
- Oroz J, Blair LJ, Zweckstetter M. (2019) Dynamic Aha1 co-chaperone binding to human Hsp90. bioRxiv https://doi.org/10.1101/550228
- Burmann BM, Hiller S. (2015) Chaperones and chaperone-substrate complexes: dynamic playgrounds for NMR spectroscopists. Prog. Nucl. Magn. Reson. Spectrosc. 86: 41-64.
- Meyer P, Prodromou C, Liao C, Hu B, Roe SM, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. (2004) Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23: 1402-1410.
- Rosenzweig R., Kay L.E. (2014) Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 83: 291-315.
- Karagoz GE, Duarte AM, Akoury E, Ippel H, Biernat J, Moran Luengo T, Radli M, Didenko T, Nordhues BA, Veprintsev DB, Dickey CA, Mandelkow E, Zweckstetter M, Boelens R, Madl T, Rudiger SG. (2014) Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 156: 963-974.
- Oroz J, Chang BJ, Wysocznski P, Lee CT, Perez-Lara A, Chakraborty P, Hofele RV, Baker JD, Blair LJ, Urlaub H, Mandelkow E, Dickey CA, Zweckstetter M. (2018) Structure and pro-toxic mechanism of the human Hsp90/PPIase/Tau complex. Nat. Commun. 9: 4523.
- Oroz J, Kim JH, Chang BJ, Zweckstetter M. (2017) Mechanistic basis for the recognition of a misfolded protein by the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 24: 407-413.
- Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, Richter K, Buchner J. (2010) Asymmetric activation of the Hsp90 dimer by its cochaperone Aha1. Mol. Cell 37: 344-354.
- Siligardi G, Hu B, Panaretou B, Piper PW, Pearl LH, Prodromou C. (2004) Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 279: 51989-51998.
doi: https://doi.org/10.1242/prelights.8801
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)