Electron Transfer from Cytochrome P450 Reductase to Cytochrome P450: Towards a Structural and Dynamic Understanding
Posted on: 27 July 2020
Preprint posted on 13 June 2020
Article now published in Communications Biology at http://dx.doi.org/10.1038/s42003-020-01568-y
Passing the electron baton: researchers use an in silico strategy to investigate electron transfer from CYP450 reductase to CYP450
Selected by Zhang-He GohCategories: bioinformatics, biophysics, pharmacology and toxicology
Background of preprint
Cytochrome P450s (CYP450s) are part of the superfamily of xenobiotic-metabolising enzymes that have been investigated for their effects on various drugs and toxins. One member of this family is CYP1A1, which is notable in two respects. First, CYP1A1 is an extrahepatic human drug target protein that modulates procarcinogen activation and carcinogen detoxification [1,2]. Second, the potential of CYP1A1 as a biocatalyst has been described as early as the last century [3,4]. Given these two essential roles of CYP1A1, a better understanding of CYP1A1 would be immensely useful to the pharmaceutical and bioengineering communities.
A two-electron transfer step between a redox partner and a heme cofactor (HEME) is often the rate-limiting step of the CYP catalytic cycle (preprint Figure 1) [5]. The mammalian CYPs have three domains (Fig. 1): a globular catalytic HEME-containing domain, an N-terminal transmembrane (TM) α-helical domain, and a flexible linker region connecting both the HEME-containing and the N-terminal TM domain. The redox partner of microsomal CYPs (including CYP1A1) is NADPH cytochrome P450 oxidoreductase (CPR). The CPR also contains three cofactor-binding domains (Fig. 1), which respectively bind to the flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and nicotinamide adenine dinucleotide (NADP).
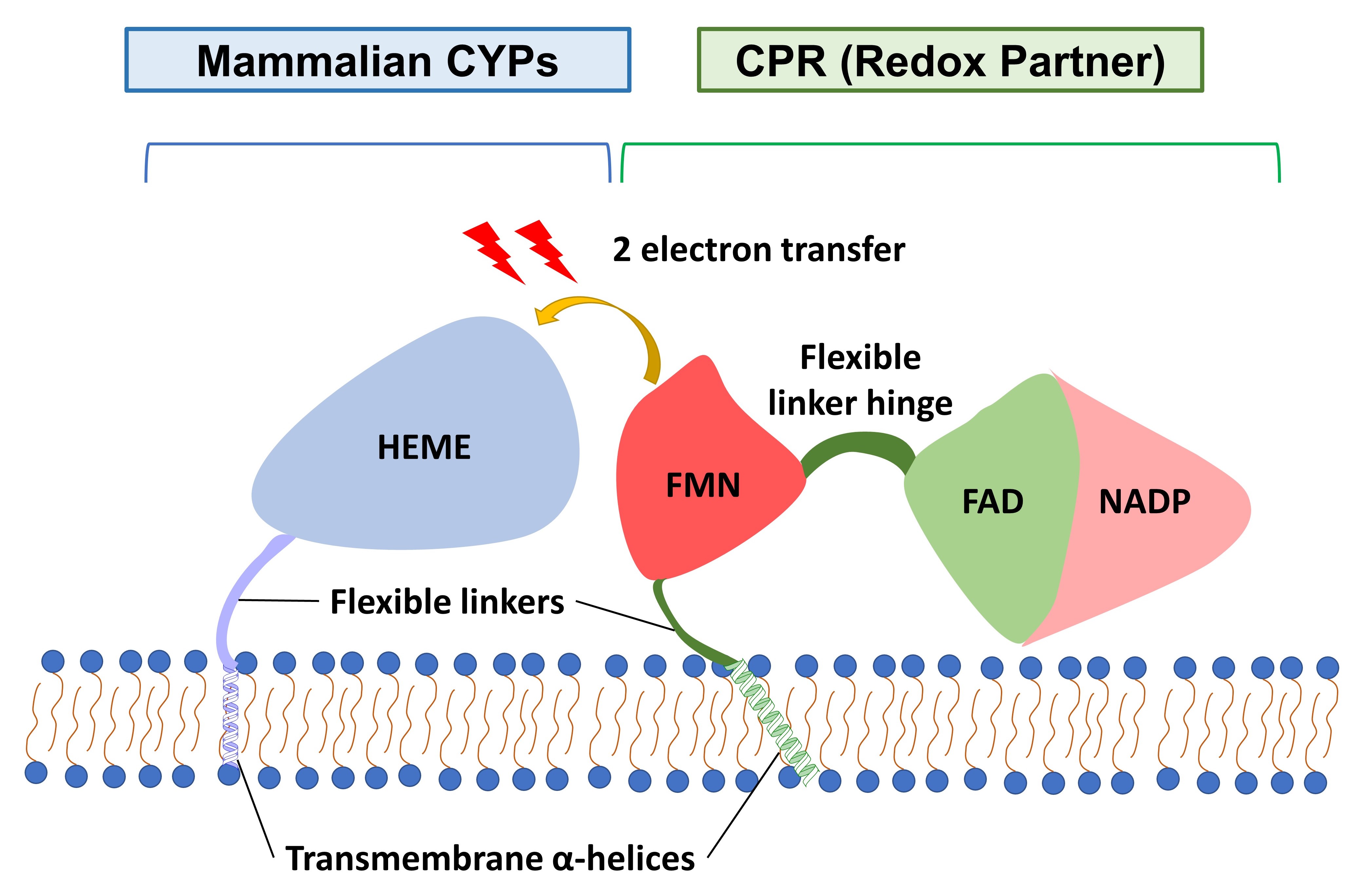
Figure 1. Mammalian CYP and CPR domains. Colours follow those in the preprint.
In their preprint, Mukherjee et al. set out to describe interactions between CYP1A1 and CPR at a detailed atomic level. They used multiresolution simulations to better understand the electron transfer process between CYP1A1 and CPR—which, in turn, informs the design of CYP1A1-targeting drugs and enhance the utility of CYP1A1 as a biocatalyst.
Key findings of preprint
Mukherjee et al. first generated a list of encounter complexes using a method known as Brownian docking (BD), an established method to predict protein-protein complexes [6]. From these, they selected six representative encounter complexes (preprint Table 1). The authors then performed structural relaxation on six encounter complexes using “soluble” molecular dynamics (MD) simulations, which are MD simulations that were conducted in aqueous solution.
After the refinement step, Mukherjee et al. narrowed their focus to just three encounter complexes simulated in the presence of a phospholipid bilayer. This led to three observations. First, the presence of CPR appeared to weaken peripheral CYP-membrane interactions. CYP1A1 became less deeply embedded in the membrane, and the CYP1A1 globular domain underwent an orientational rearrangement. Second, the FMN domain of CPR rearranged with respect to the membrane, largely caused by rearrangements of the CPR’s highly flexible linker region. Third, the FAD and NADP domains also rearranged with respect to the membrane. This made the CPR structure more compact, adopting a conformation resembling the semi-open one observed in the crystal structure.
Based on the high contact area between CYP1A1 and CPR, the authors concluded that CYP1A1 and CPR formed strongly-bound and stable transient complexes in the membrane. The CYP-FMN domain interactions involved the pairing of the positively-charged CYP1A1 proximal side with the negatively-charged FMN domain; such charge pairings have been identified as important contributors towards redox protein binding. In contrast, transient interactions formed between CYP1A1 and the NADP domain of the CPR. The authors identified two major electron transfer pathways, FMN-I548-C457-HEME and FMN-K456-C457-HEME; both corroborate other experimental data [7,8] and computational studies [9-11].
Finally, Mukherjee et al. also investigated the effects of CPR binding on the opening of ligand tunnels between the buried CYP active site and the protein surface. Specifically, the authors used conventional MD simulations, as well as random acceleration MD (RAMD) simulations of ligand egress from the CYP active site. While the authors could not draw causal inferences between CPR binding and changes in the conformations of the ligand tunnels, their RAMD simulations showed that the binding of CPR to CYP1A1 alters the distribution of egress pathways. This latter finding supports MD simulations in a recent study that CPR binding modulates ligand tunnels [12].
What I like about this preprint
In their preprint, Mukherjee et al. described their findings from a computational approach to characterising the electron transfer between CYP1A1 and CPR, its redox partner. The authors’ in silico strategy revealed key interactions between the two partners, as well as their subsequent conformational changes, that facilitate electron transfer.
The in silico approach adopted by Mukherjee et al. reinforces current experimental and computational findings on CYP450 enzymes reported by other groups. Their findings are especially pertinent when we consider key mutations that may affect the activity of these enzymes, which are implicated in both xenobiotic metabolism [7] and in certain cancers [13,14].
Future work
These insights made in CYP1A1 will enhance researchers’ understanding of the CYP450 landscape. A mechanistic investigation of the impact of CYP450 conformational structure on its kinetics may help bioengineers to optimise processes using these enzymes in synthetic biology, allowing them to synthesise complex molecules more efficiently.
The authors’ findings will also help pharmaceutical developers to design better drugs. By elucidating the effect of CYP-CPR binding on ligand binding—as well as substrate access and product release—in their preprint, Mukherjee et al. open new avenues for drug design and development. For one, anticancer drugs may be developed by targeting CYP1A1, which has been implicated in cancer aetiology. Alternatively, the headway made into CYP1A1 may help pharmacokineticists better predict drug-drug interactions involving CYP enzymes.
Like the electron cascade it described, this preprint marks the start of new inroads made in enzymatic discovery. It will be electrifying to see where it goes next.
References
[1] Androutsopoulos VP, Tsatsakis AM, Spandidos DA, Cytochrome P450 CYP1A1: wider roles in cancer progression and prevention, BMC Cancer 9(1) (2009) 187.
[2] Nandekar PP, Sangamwar AT, Cytochrome P450 1A1-mediated anticancer drug discovery: in silico findings, Expert Opin Drug Discov 7(9) (2012) 771-789.
[3] Sakaki T, Kominami S, Takemori S, Ohkawa H, Akiyoshi-Shibata M, Yabusaki Y, Kinetic studies on a genetically engineered fused enzyme between rat cytochrome P4501A1 and yeast NADPH-P450 reductase, Biochemistry 33(16) (1994) 4933-4939.
[4] Murakami H, Yabusaki Y, Sakaki T, Shibata M, Ohkawa H, A genetically engineered P450 monooxygenase: construction of the functional fused enzyme between rat cytochrome P450c and NADPH-cytochrome P450 reductase, DNA 6(3) (1987) 189-197.
[5] Guengerich FP, Mechanisms of cytochrome P450 substrate oxidation: MiniReview, J Biochem Mol Toxicol 21(4) (2007) 163-168.
[6] Motiejunas D, Gabdoulline R, Wang T, Feldman-Salit A, Johann T, Winn PJ, Wade RC, Protein-protein docking by simulating the process of association subject to biochemical constraints, Proteins 71(4) (2008) 1955-1969.
[7] Lewis BC, Mackenzie PI, Miners JO, Application of homology modeling to generate CYP1A1 mutants with enhanced activation of the cancer chemotherapeutic prodrug dacarbazine, Mol Pharmacol 80(5) (2011) 879-888.
[8] Fernández-Cancio M, Camats N, Flück CE, Zalewski A, Dick B, Frey BM, Monné R, Torán N, Audí L, Pandey AV, Mechanism of the Dual Activities of Human CYP17A1 and Binding to Anti-Prostate Cancer Drug Abiraterone Revealed by a Novel V366M Mutation Causing 17,20 Lyase Deficiency, Pharmaceuticals (Basel) 11(2) (2018).
[9] Šrejber M, Navrátilová V, Paloncýová M, Bazgier V, Berka K, Anzenbacher P, Otyepka M, Membrane-attached mammalian cytochromes P450: An overview of the membrane’s effects on structure, drug binding, and interactions with redox partners, Journal of Inorganic Biochemistry 183 (2018) 117-136.
[10] Ritacco I, Spinello A, Ippoliti E, Magistrato A, Post-Translational Regulation of CYP450s Metabolism As Revealed by All-Atoms Simulations of the Aromatase Enzyme, Journal of Chemical Information and Modeling 59(6) (2019) 2930-2940.
[11] Fishelovitch D, Hazan C, Hirao H, Wolfson HJ, Nussinov R, Shaik S, QM/MM study of the active species of the human cytochrome P450 3A4, and the influence thereof of the multiple substrate binding, J Phys Chem B 111(49) (2007) 13822-13832.
[12] Ritacco I, Saltalamacchia A, Spinello A, Ippoliti E, Magistrato A, All-Atom Simulations Disclose How Cytochrome Reductase Reshapes the Substrate Access/Egress Routes of Its Partner CYP450s, The Journal of Physical Chemistry Letters 11(4) (2020) 1189-1193.
[13] Freedland J, Cera C, Fasullo M, CYP1A1 I462V polymorphism is associated with reduced genotoxicity in yeast despite positive association with increased cancer risk, Mutat Res 815 (2017) 35-43.
[14] Ezzeldin N, El-Lebedy D, Darwish A, El-Bastawisy A, Hassan M, Abd El-Aziz S, Abdel-Hamid M, Saad-Hussein A, Genetic polymorphisms of human cytochrome P450 CYP1A1 in an Egyptian population and tobacco-induced lung cancer, Genes Environ 39 (2017) 7-7.
Acknowledgements
The preLight author is grateful to the preprint authors for their suggestions regarding Figure 1.
doi: https://doi.org/10.1242/prelights.22995
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the pharmacology and toxicology category:
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
In vitro pharmacokinetics and pharmacodynamics of the diarylquinoline TBAJ-587 and its metabolites against Mycobacterium tuberculosis
Zhang-He Goh
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the pharmacology and toxicology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Drug use in special populations
Any drugs that are being used in special populations: Patients with liver and kidney failure, in paediatrics, in geriatrics, and in pregnant or lactating patients. Includes the discovery of factors that could potentially affect drug use in these special populations.
| List by | Zhang-He Goh |
Toxicology of toxicants, existing therapeutics, and investigational drugs
Preprints that describe the toxicology of environmental pollutants and existing and upcoming drugs. Includes both toxicokinetics and toxicodynamics, as well as technological improvements that will help in the characterisation of this field.
| List by | Zhang-He Goh |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)