Endogenous retroviruses drive species-specific germline transcriptomes in mammals
Posted on: 27 May 2020
Preprint posted on 11 March 2020
Article now published in Nature Structural & Molecular Biology at http://dx.doi.org/10.1038/s41594-020-0487-4
A case of endogenous retroviral control of male germline genes in mammals
Selected by Petra KovacikovaCategories: developmental biology, evolutionary biology
Background:
Transposable elements (TE) are sequences of DNA capable of changing their position in the genome by first being cut from their initial position and then pasted in a new place. They occupy a surprisingly large portion of mammalian genomes and have long been thought to be just “junk” DNA, often lacking coding potential for any protein except the transposase which is responsible for the jumping feature of these genes. Some of TEs are derived from endogenous retroviruses (ERV), retroviruses that integrated into the germline genome and have been inherited for generations. In fact, many have lost the ability to transpose completely. Nonetheless, they pose a threat to the host genome by introducing mutations in place of their integration, hence the cell tries to prevent such events by methylation or other silencing mechanisms of the chromatin region they lie in. This is especially true for germ cells that carry the heritable genome.
In this preprint, the authors envisage the possible mechanism for germline transcriptome divergence in mammals by looking at the role of endogenous retroviruses (ERV) in the male germline. Despite their presumably deleterious effects on the germline, this class of transposable elements is known to regulate germline expression patterns, mainly through influencing post‑transcriptional control or promoter function. This time, however, the authors present evidence for the evolutionarily young K family of endogenous retroviruses (ERVKs) being used as cis-regulatory elements, containing binding sites for the master regulator of male meiosis and driving transcription of genes important for the mitosis-meiosis transition in the germline.
Key findings:
Endogenous retroviruses are differentially expressed at the mitosis-meiosis transition in the male germline, reside in open chromatin, exhibit enhancer-like histone marks and bind a key spermatogenesis master regulator
To test the hypothesis whether ERVs contribute to dynamic transcriptomic changes during spermatogenesis, the authors analyzed published RNA-seq datasets from 5 representative stages of mouse spermatogenesis from postnatal day 7 to adulthood (Fig.1 a).
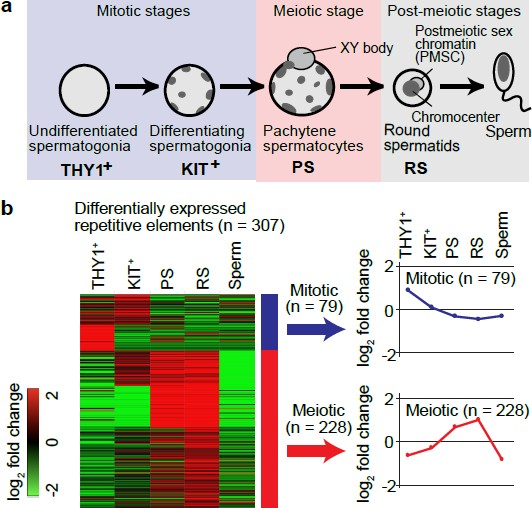
Differential expression of mouse consensus repetitive elements revealed two temporally regulated groups of ERVS: mitotic and meiotic (Fig.1 b). The most represented types of TEs in both groups were members of ERVK, ERV1 and ERVL families of endogenous retroviruses. Interestingly, all of them were enriched in intergenic or intronic regions of the mouse genome and didn’t have any significant coding potential. Genome-wide chromatin accessibility assays showed that only some part of ERV loci exhibit open chromatin. Clustering of accessible ERV loci across different timepoints in spermatogenesis again resulted in mitotic and meiotic groups. The most represented family of meiotic class, ERVK, showed strong enrichment for three subgroups: RLTR10, RMER17 and RLTR51. Likewise, the enrichment of ATAC- peaks for these subgroups turned out to be specific for late spermatogenesis, based on the comparison to ATAC- seq data from cell lines derived from other tissues. In addition to tissue specificity, many of accessible RLTR10 and RMER17 in pachytene stage mapped to sex chromosomes compared to autosomes. This would mean that they become active despite ongoing meiotic sex chromosome inactivation at this meiotic phase.
Even more telling about the function of these ERVK open chromatin loci is the fact that they carry the histone modification H3K27ac, typical of enhancer sequences. Enhancer-like function is further supported by the presence of the binding site for A-MYB, a key transcription factor that acts as a master regulator of male germline from early meiosis. Chromatin immunoprecipitation with A-MYB antibodies recovers the same loci as ERVK enhancer-like loci on both autosomes and sex chromosomes. A significant portion of genes located in ERVK enhancer-like loci proximity are highly
expressed in meiosis and are involved in processes like chromosome segregation, chromatin silencing and spermatogenesis. More importantly, some of these genes are differentially expressed in A-MYB mutant mouse, indicating that A-MYB is indeed the mediator of ERVK enhancer-like activation of transcription.
Evolutionary conservation of ERVs driving species-specific spermatogenesis needs?
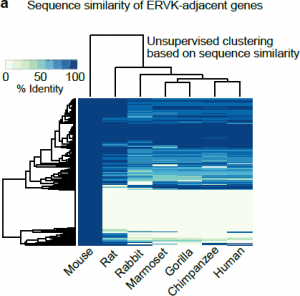
Perhaps even more surprising is the orthology assignment of these ERVK-adjacent genes. It seems, that for the most part, they consist of evolutionary young genes that share very little sequence identity (Fig 2), even if compared to the rat within the same clade. How frequent then is this phenomenon of ERV-based expression regulation in evolution? Analysis of human testis expression and immunoprecipitation datasets pointed at members of ERV1 and ERVK families, bearing binding motifs for A-MYB, as possible enhancers of mitosis-meiosis transition genes. This led the authors to propose that ERVK-driven meiotic enhancers are a general feature of mammals responsible for divergence of transcriptomes in late spermatogenesis.
Why is this work important?
The work of Sakashita and colleagues nicely demonstrates the innovative use of repetitive elements in genome regulation. While it builds on previous studies that focused on the function of endogenous retroviruses in male meiosis, it offers a thought-provoking view on the possible involvement of retroviral elements in an important task such as forming a species-specific germline transcriptome and eventually the gametes. I enjoyed this different take on underlying mechanisms of controlling dynamics of the germline transcription landscape, while exploring the avenue of non-coding repetitive elements’ purpose that we still know very little about.
Questions for the authors:
- Did the authors look into differential expression of repetitive elements in oogenesis? Would they expect to see the same trend with sharp difference between mitotic and meiotic clusters?
- Could the authors describe their algorithm of k-means clustering of ATAC-peaks (used for Fig 2a) in more detail? Did the selection of k=2 come out as the most suitable option for the analysis, or were there any subtle changes caused by increasing the k value that would point to a finer and/or gradual regulation rather than a very “sharp” line on mitosis-meiosis transition?
- Approximately one-fifth of the ERVK-adjacent genes involved in mitosis-meiosis transition are under control of A-MYB transcription factor (results of A-MYB mutant mouse data analysis). Do the authors have any hypothesis on which potential TFs could bind to ERVK enhancer-like loci in proximity of the remaining ERVK-adjacent genes?
- As the authors outlined in the discussion, one possibility of how the accessibility of ERVK enhancers loci chromatin is being so tightly regulated at the mitosis-meiosis boundary is by the function of KRAB domain zinc finger proteins. However, the ATAC-seq data shows that in pre meiotic period, the chromatin of “meiotic” ERVK loci is closed. Could it then be simply inaccessibility that prevents A-MYB from binding rather than suppression by KRAB-ZF?
- Looking at the Figure 5a in the paper (Fig2 in here), it seems there is a large fraction of ERVK-adjacent genes (Supplemental table 2) with sequence identity >70 % (upper cluster in the Fig 5a). Are these the genes associated with the GO terms “chromosome segregation” and “chromatin silencing”, (or possibly some of the other enriched GO terms) rather than spermatogenesis? Perhaps, it would be more evident for the reader to distinguish which subset of these genes the text and conclusions refer to if the clusters were labelled (also in the Suppl. Table2 which could include the GO terms?). However, the overall conclusion in the paper is that the ERVK-adjacent genes are not conserved in closely related species, let alone other mammals (“no unambiguous homolog found”). Is it the portion of A‑MYB dependent genes that is species-specific and forms the basis for this conclusion? If so, did the authors try to explore the genomic environment of ERVK loci in other mammalian species for which spermatogenesis ATAC‑seq data are available?
- How likely do the authors think this phenomenon of using ERVs as gametogenesis specific enhancers can be found outside of mammals? Or perhaps even in invertebrates?
doi: https://doi.org/10.1242/prelights.21142
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
Also in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)