Establishment of the mayfly Cloeon dipterum as a new model system to investigate insect evolution
Preprint posted on 12 December 2018 https://www.biorxiv.org/content/10.1101/494674v1
Article now published in EvoDevo at http://dx.doi.org/10.1186/s13227-019-0120-y
Filling the phylogenetic gap in insect evo-devo: The Mayfly Room, and the establishment of an evolutionary ancient group of insects in the lab
Selected by Ivan Candido-FerreiraBackground
Insects represent one of the most evolutionarily successful radiations of terrestrial animals dating back from more than 490 million years ago. However, since the early establishment of fruit flies (Drosophila melanogaster) as an experimental system, an event pivotal for modern genetics and molecular biology, tractable systems for evolutionary developmental studies within insects have been largely limited to species that represent very derived lineages within the insect phylogeny. In other words, the evolutionary history of insects has not been taken into account when model systems were established in the lab, mainly owing to practical limitations such as culturing methods and maintenance costs. Such scarcity of diverse insect systems tractable in a lab has thus negative implications for our current understanding of the key events underlying the evolution of insect novelties, such as wings, during their early radiation.
Among other groups of animals, this limitation has been tackled by establishing culture systems of taxonomically diverse groups, with species that represent lineages strategically positioned within their phylogenies. For example, the establishment of lampreys, an extant group of basal vertebrates, and of urochordates such as the sea squirt Ciona (which belongs to a group that represent the sister lineage of vertebrates), as an alternative to evolutionary, developmental and genomics studies largely conducted in humans, mice, chicken, frogs and zebrafish, have been key to a better understanding of the transitions that enabled the evolution and diversification of vertebrates.
It is worth noting, however, that such attempts to establish phylogenetically well-positioned systems can be deemed heroic, sometimes. For example, in addition to lampreys, hagfishes are the only extant jawless vertebrates that could be used to study the developmental and molecular biology of basal vertebrates. However, the majority of hagfish species are inhabitants of the deep ocean, and it took more than 100 years to establish the first culture of hagfishes in a lab that yielded embryos that comprehensively allowed the study of hagfish early development.
The preprint
In a new preprint, Isabel Almudi and colleagues at the Casares lab in the CABD (Sevilla, Spain), established the first continuous culture of mayflies (Figure 1), and also provided a wealth of transcriptomic data. Together with dragonflies (Odonata), mayflies belong to the most ancient group of winged insects, the Paleoptera.

Figure 1. An adult male of Cloeon dipterum (Ephemeroptera: Baetidae). Photo courtesy of Isabel Almudi. Notice the extra pair of very large dorsal eyes, called the turbanate eyes – a striking sexual dimorphism in mayflies from the Baetidae family.
However, all insects used in evolutionary developmental biology and genomics studies are positioned within a group of highly derived lineages, the holometabolans, including the fruit fly Drosophila, the beetle Tribolium and Heliconius butterflies. Therefore, the ancient placement of mayflies in the insect phylogeny allows for the interrogation of the early evolution of insects. In other words, mayflies represent to Drosophila what lampreys represent to humans. Almudi et al ‘s work is thus poised to illuminate the early transitions during winged insects’ evolution and test postulated hypothesis for wing evolution.
The model system
Mayflies have two key distinct phases in their life cycle, the aquatic, which represents the nymphal stages, and the terrestrial, which represents the post-metamorphic (pre-imago) and the reproductive stage (imago). Mayflies also possess a remarkable diversity of adaptations, and their order (Ephemeroptera) is unique in that after the last moult the reproductive adults aren’t capable of feeding due to the presence of vestigial mouthparts, and thus die not long after mating (hence their order name, ephemera = short-lived).
Almudi et al used an ovoviviparous species of mayfly, C. dipterum, collected in Southern Spain. Lines were established from single gravid females, which layed the eggs in a Petri dish. Usually mayflies mate during the flight, so forced mating (Figure 2) was attempted and spawning was induced within days until fertile eggs were layed (varying in clutches of 100-300 eggs, depending on the nutritional state of the female). In fact, such ease of pairing individuals represents a useful framework in which lines with interesting phenotypes/genotypes can be established. Future development of RNAi and/or CRISPR/Cas9 techniques in this system will be useful to test this.

Figure 2 – Mating of C. dipterum in the lab. Top: female, bottom: male. From the preprint Figure 2a.
Importantly, males and females are easily distinguishable, and nymphs feed on algae. Males are discarded after mating, and the induced spawning of females allows for a highly synchronised, high number of embryos being obtained for multiple experiments.
Establishment of the lines allowed Almudi et al to comprehensively survey the entire embryogenesis of C. dipterum, which takes around 15 days to complete. Moreover, transcriptomic data from a single male nymph allowed the de novo reconstruction of 117233 transcripts (Figure 3), a resource that will not only be useful for the design of riboprobes, RNAi and single RNA guides for CRISPR/Cas9 studies, but also for future phylogenomic studies. Ongoing work to sequence the C. dipterum genome, which consist in a karyotype of 2N=10, will help to foster the study of this emerging system in evolutionary developmental biology.
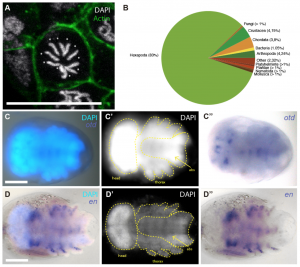
Figure 3 – Transcriptomics in C. dipterum. (a) The karyotype of C. dipterum (2N=10), (b) transcriptome homology to other organisms, (c-d) In situ hybridization for orthodenticle and engrailed transcripts. From the preprint Figure 5.
Regenerative capacities of a mayfly
Almudi et al also push forward their system by showing preliminary data regarding the exceptional regenerative capacities of post-embryonic stages of C. dipterum. In fact, within 7 days nymphs are fully capable of regenerating a full leg (Figure 4), in contrast to one month in crickets and 18+ weeks in cockroaches. Therefore, C. dipterum could emerge as an attractive model owing to its extraordinarily fast limb regeneration capacities in post-embryonic stages.

Figure 4. Full leg regeneration in a C. dipterum nymph within 7 days. From the preprint Figure 6.
Conclusions
Despite the key importance of insects in agriculture, ecology, genetics and physiology, insect molecular biology has largely been restricted to the fruit fly Drosophila melanogaster, and is only recently expanding to other holometabolan insects, such as Tribolium and Heliconius.
In this preprint, Almudi et al were able to move the field of insect evolutionary developmental biology forward by establishing a continuous culture of an insect that belongs to an ancient group, the Paleoptera. Such system will allow the comprehensive interrogation of testable hypothesis within the embryogenesis and post-embryonic development of a mayfly, such as the role of homeotic transformations in the early appearance of insect wings. Therefore, their system is strategically well positioned for testing postulated hypotheses about the evolution and diversification of insects, as well as emergent properties such as the extraordinary regenerative capacities of C. dipterum. In the very style of tackling old questions proposed by Prof. Alejandro Sanchez-Alvarado, Almudi et al’s work thus aims to resolve “old questions by studying new species”.
Questions to the authors
- What were the key obstacles in establishing C. dipterum in the lab? For example, how did you realise forced mating was going to be necessary, and how hard was it to achieve a successful mating in the first time?
- What reasons led you to choose to culture a mayfly and not other paleopteran, such as dragonflies?
- At what stage are the eggs when they are laid? If mostly single celled, is it feasible to inject/electroporate constructs, or mosaicity is going to be a challenge to tackle in transgenic lines?
- The regenerative capacities you highlight in the preprint seem to be really extraordinary. Are there similar capacities in sub-imago adults? Do you have any evidence to say whether, mechanistically, regeneration is similar as to Parhyhale? Could it be related to mounting in nymphs?
Related References
Dewitz, H. 1890. Zool. Anz. 13, 535
Ota, K. G., Kuraku, S. & Kuratani, S. Hagfish embryology with reference to the evolution of the neural crest. Nature 446, 672–675 (2007).
Misof, B. et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767 (2014)
The Fly Room (2014), directed by Alexis Gambis
Prof. Alejandro Sánchez Alvarado TED talk “To solve old problems, study new species” (2017), available online at https://www.ted.com/talks/alejandro_sanchez_alvarado_to_solve_old_problems_study_new_species
Posted on: 21 February 2019 , updated on: 22 February 2019
doi: https://doi.org/10.1242/prelights.8879
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the animal behavior and cognition category:
Experience-dependent plasticity of a highly specific olfactory circuit in Drosophila melanogaster
Behavioral screening of conserved RNA-binding proteins reveals CEY-1/YBX RNA-binding protein dysfunction leads to impairments in memory and cognition
Pervasive sublethal effects of agrochemicals as contributing factors to insect decline
Also in the developmental biology category:
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Also in the ecology category:
How the liver contributes to stomach warming in the endothermic white shark Carcharodon carcharias
Pervasive sublethal effects of agrochemicals as contributing factors to insect decline
Multimodal interactions in Stomoxys navigation reveals synergy between olfaction and vision
Also in the evolutionary biology category:
Fetal brain response to maternal inflammation requires microglia
How the liver contributes to stomach warming in the endothermic white shark Carcharodon carcharias
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
Also in the zoology category:
How the liver contributes to stomach warming in the endothermic white shark Carcharodon carcharias
Unlocking the secrets of kangaroo locomotor energetics: Postural adaptations underpin increased tendon stress in hopping kangaroos
Multimodal interactions in Stomoxys navigation reveals synergy between olfaction and vision
preLists in the animal behavior and cognition category:
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Also in the developmental biology category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the ecology category:
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
Also in the evolutionary biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (2 votes)
(2 votes)