Hedgehog signaling can enhance glycolytic ATP production in the Drosophila wing disc
Posted on: 8 October 2021 , updated on: 19 December 2022
Preprint posted on 12 September 2021
Article now published in EMBO reports at http://dx.doi.org/10.15252/embr.202154025
Hedgehog and glycolysis: the perfect team to regulate energy production in the developing Drosophila wing disc.
Selected by Julia GrzymkowskiCategories: developmental biology, physiology
Updated 18 October 2022 with a postLight by Julia Grzymkowski
In looking at both the preprint of this manuscript and the final version, which was recently published in EMBO Reports (http://dx.doi.org/10.15252/embr.202154025), there were some changes and additions made to the manuscript which have increased the robustness of the study and validity of the conclusions.
Some small changes were made to Figure 1. Specifically, another time point was added to the antimycin A treatment experiments, which tightens the quantitative data. The difference in the Hill coefficient that could be observed in the data included in the preprint is no longer there in the data of the final manuscript, but this has not changed the overall conclusions. Given that the half-life of the FRET efficiency was longer in the organizer regions, the data still indicates that ATP levels decline at a slower rate in the organizer regions of the Drosophila wing disc compared to non-organizer regions.
Later in the manuscript, the authors decided to combine the original Figures 3 and 4 into one figure (now Figure 3) in the final manuscript and then added a new set of experiments for their new Figure 4. These set of experiments looked at the effects of a hypoxic environment on ATP levels in the wing disc. They found similar spatial differences in ATP levels upon decreased levels of oxygen as compared to OxPhos inhibition. These experiments replicate the realistic hypoxic conditions that can be observed during normal Drosophila larval development.
While the final published version of the paper does not address the questions posed in my preLight, this does not take anything away from it. Those questions were outside the scope of the manuscript and may be discussed in a later study.
Background
Developmental tissue growth is an energy-dependent process that relies both on ATP levels and the production of biosynthetic precursors. The concept of “aerobic metabolism” (i.e., Warburg effect), where increased glucose uptake occurs despite active respiration, has been widely studied in the context of cancer. The increased glucose uptake allows cancer cells to redirect glycolytic metabolites to biosynthetic pathways needed for growth. Recently, studies have emerged showing that some developing tissues and/or cells display aerobic metabolism as well (1-3), and that glycolysis regulates developmental processes such as migration and differentiation. In addition, connections between morphogen signaling (e.g., Wnt, FGF, Hh) and metabolism were established lately in different developmental contexts, and it is of continued interest to determine how manipulating one may affect the other (1, 4). However, research in this field is in its infancy. In their preprint, Nellas et al use a fluorescent biosensor to study the spatial dynamics of ATP within the Drosophila wing disc and how Hedgehog (Hh) signaling influences energy production during development of this tissue.
Key Findings
Using a FRET-based ATP sensor, the authors found that ATP levels remain steady throughout the wing pouch, but decrease significantly upon oxidative phosphorylation (OxPhos) inhibition with antimycin A. Upon closer inspection, the authors noticed that with antimycin A treatment, ATP levels declined significantly slower in two specific regions of the wing disc as compared to the rest. These regions of slower ATP decline were within known growth “organizer” regions: in the Dorsal-Ventral (DV) boundary, which has high Wingless/Notch signaling, and near the Anterior-Posterior (AP) boundary, which has high Hh signaling. The authors posited that these organizer regions may produce more ATP through glycolysis during OxPhos inhibition, and that this may explain why ATP levels decline more slowly. Inhibiting both OxPhos and glycolysis concomitantly resulted in a steep decline in ATP levels throughout the entire wing disc, including the organizer regions. Therefore, the authors concluded that most ATP in the wing disc is generated by OxPhos, but when OxPhos is inhibited, glycolysis continues to generate significant levels of ATP within the organizer regions.
These results suggest that morphogen signaling may control energy production within these growth organizer regions. Hh is expressed in the posterior compartment of the wing disc, where it then travels to the anterior to be received by the membrane protein, Patched (Ptc). Ptc is normally a Hh signaling repressor in the absence of Hh ligand, where binding of Hh to Ptc allows activation of gene expression through the downstream transcription factor Cubitus interruptus (Ci). Ptc was inhibited within the dorsal compartment of the wing disc via RNAi, leaving the ventral compartment as an internal control (Fig. 1A). This resulted in an overactivation of the Hh pathway within the dorsal anterior region, which had a significantly slower decline of ATP levels after OxPhos inhibition (Fig. 1C). To further assess whether the Hh pathway is required for glycolysis in the wing disc, a dominant negative form of Ci was overexpressed within the dorsal compartment. Upon loss of Hh signaling in this region, ATP levels declined faster than within the ventral compartment after treatment with antimycin A. Altogether, the results presented here and in their previous work (4), reveal a positive feedback loop between Hh and glycolysis that is needed to couple energy production and patterning during development.
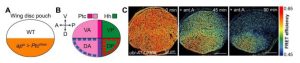
Figure 1. A) Ptc is inhibited only in the dorsal compartment of the wing disc, resulting in an overactivation of the Hh pathway in the dorsal anterior (DA) region (B). C) Increased Hh signaling leads to slower ATP depletion in the DA region of the wing disc after antimycin A treatment. (Adapted from Figure 3, Nellas et al. 2021)
Why I chose this preprint
This preprint utilizes a simple but powerful system, the Drosophila wing disc, to further study the interplay between metabolism and morphogen signaling, a topic where little is known. The results presented here add important and convincing evidence that developmental patterning events rely on metabolism, and vice versa. In addition, utilization of the FRET-based ATP sensor offers spatial information that is crucial when studying patterning events. Studies like this are helping to lay the foundation for other researchers exploring metabolic regulation during development.
Questions for the authors
- When assessing the kinetics of ATP depletion, in some instances the Half-life would show a significant difference between regions, whereas the Hill coefficient was not significantly different (e.g., Fig 3). Could the authors comment on whether there is a difference in reliability of readouts of kinetics?
- It looks like the dorsal portion of the wing disc that is expressing the CiDN (and therefore has reduced Hh signaling/glycolysis) is visibly smaller than the ventral WT portion of the wing disc. Did you see any abnormal outgrowth/increased growth within the dorsal anterior region of the PtcRNAi wing disc (which has increased Hh signaling/glycolysis)?
- A limited number of studies have made a connection between NAD+ metabolism and morphogen signaling (5,6). Considering the importance of NAD+ in glycolysis and OxPhos, do you think NAD+ levels could have an influence on Hh signaling in your system?
References:
- Oginuma, M., Moncuquet, P., Xiong, F., Karoly, E., Chal, J., Guevorkian, K., & Pourquié, O. (2017). A Gradient of Glycolytic Activity Coordinates FGF and Wnt Signaling during Elongation of the Body Axis in Amniote Embryos. Developmental Cell, 40(4), 342–353.e10.
- Bulusu, V., Prior, N., Snaebjornsson, M. T., Kuehne, A., Sonnen, K. F., Kress, J., Stein, F., Schultz, C., Sauer, U., & Aulehla, A. (2017). Spatiotemporal Analysis of a Glycolytic Activity Gradient Linked to Mouse Embryo Mesoderm Development. Developmental Cell, 40(4), 331–341.e4.
- Bhattacharya, D., Azambuja, A. P., & Simoes-Costa, M. (2020). Metabolic Reprogramming Promotes Neural Crest Migration via Yap/Tead Signaling. Developmental Cell, 53(2), 199–211.e6.
- Spannl, S. et al. (2020). Glycolysis regulates Hedgehog signaling via the plasma membrane potential. The EMBO Journal, 39(21), p. e101767. doi: 10.15252/embj.2019101767.
- Piao, J., Tsuji, K., Ochi, H. et al. (2013). Sirt6 regulates postnatal growth plate differentiation and proliferation via Ihh signaling. Sci Rep, 3, 3022. https://doi.org/10.1038/srep03022.
- Lee, J., Kee, H. J., Min, S. et al. (2016). Integrated omics-analysis reveals Wnt-mediated NAD+ metabolic reprogramming in cancer stem-like cells. Oncotarget, 7(30), 48562–48576. https://doi.org/10.18632/oncotarget.10432
doi: https://doi.org/10.1242/prelights.30782
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
Also in the physiology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the physiology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |











 (1 votes)
(1 votes)