Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene
Posted on: 10 July 2018 , updated on: 11 July 2018
Preprint posted on 11 June 2018
Article now published in eLife at http://dx.doi.org/10.7554/eLife.61635
Falling into lines: @guberrocal, @HernanGGarcia and @mbeisen use live imaging and computational modelling to reveal the transcriptional dynamics underlying the emergence of the eve stripes in the Drosophila embryo
Selected by Erik ClarkCategories: biophysics, developmental biology
Background
Segmentation gene expression in the Drosophila blastoderm has long been used as a test case for unravelling mechanisms of transcriptional regulation. Over the years, one of the most scrutinised genes has been even-skipped (eve), a pair-rule gene expressed in seven regular stripes. Although it was originally assumed that the regularity of these stripes must stem from an underlying uniformity of mechanism (e.g. Turing pattern formation), it was quickly discovered that the various eve stripes are actually established piecemeal, from 5 different “stripe-specific” enhancer elements. Today, these enhancers are among the best studied in the animal kingdom.
In recent years, the MS2-MCP system for live imaging has shone a light on the dynamicity of transcriptional regulation, again using Drosophila blastoderm genes such as eve as study cases. Transgenically inserting MS2 stem-loop repeats into a coding sequence causes maternally deposited and fluorescently tagged MCP protein to be concentrated at the gene locus during active transcription, allowing one to directly visualise transcriptional bursts in individual nuclei over time. However, in order to determine how exactly transcription is regulated across time and space to pattern developmental gene expression, it is first necessary to overcome the problem of inferring the promoter state history of each nucleus from its temporal intensity profile (an aggregate pattern which is determined by the cumulative number of nascent transcripts present at the transcribing locus over time).
Key Findings
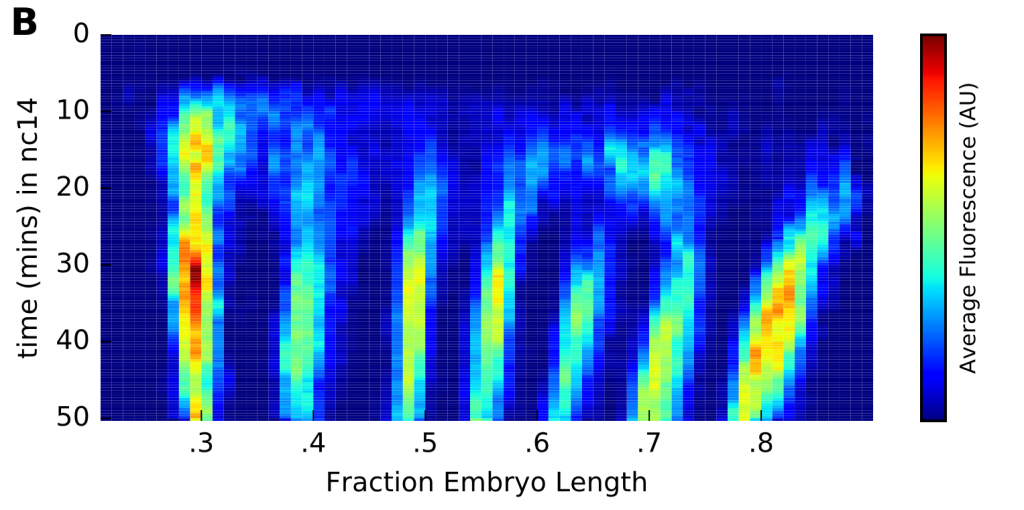
Berroccal and colleagues constructed an eve MS2 reporter transgene, and collected 11 high-resolution movies of blastoderm eve expression, each focused on a different set of stripes. They segmented the movies to get the locations and intensity profiles of ~3,000 individual nuclei, then registered all this data together to produce an overall picture of eve transcription along the anteroposterior axis during and after stripe formation.
The authors then ran the intensity traces through a custom hidden Markov model to infer the history of transcriptional bursts in each nucleus (see the companion preprint in Related Research, below). A simple two-state (ON/OFF) modelling framework is commonly used to characterise transcriptional regulation at the promoter level, characterised by three parameters, which determine (1) average burst frequency, (2) average burst duration, and (3) the rate at which new transcripts are initialised during a burst. The authors found that eve transcription is regulated predominantly at the level of burst frequency (increasing it within stripes, and decreasing it between stripes), while the other two parameter values did not seem to change much. They also found that the different eve stripes are regulated similarly at the promoter level, even though they are generated by different enhancer elements and patterned by different sets of transcription factors.
Reassuringly, the tissue-level picture of eve expression yielded by the analysis was broadly in line with previous quantitative descriptions derived from large numbers of fixed samples. However, the authors were able to put more precise timings on the resolution of individual stripes, and more accurately determine the magnitude of the posterior to anterior shifts undertaken by the eve stripes after they form. Overall, this study underlines the extremely dynamic nature of eve expression within the blastoderm – as the authors stress, “at no point does eve approach anything even remotely like a steady state”.
Significance
We are going to see many more MS2 movies in developmental biology over the coming years. This windfall of live imaging data will need to be analysed rigorously, perhaps using a similar framework to the one laid out here. This preprint therefore represents an important technical development. It also represents a laudable example of Open Science best practise: all datasets are well-documented and available in an online labbook, and Michael Eisen even wrote a great explainer thread for the project on Twitter: https://twitter.com/mbeisen/status/1002430064225480704
Recently, Hernan Garcia and colleagues revealed a complementary live imaging technology (LlamaTags), which allows live imaging of transcription factors. This means that it is now possible to analyse both halves of the equation: we can characterise transcriptional dynamics at the single cell level, as seen in this preprint, but also relate these outputs to the changing concentrations of key regulatory proteins within these same cells. It seems that old friends like even-skipped will have much to teach us for many years to come.
Related Research
Lammers NC, Galstyan V, Reimer A, Medin SA, Wiggins CH, Garcia HG (2018) Binary transcriptional control of pattern formation in development. bioRxiv doi: 10.1101/335919
Bothma JP, Norstad MR, Alamos S, Garcia HG (2018) LlamaTags: a versatile tool to image transcription factor dynamics in live embryos. Cell 173 1810-22
Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M (2014) Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. PNAS 111 10598-10603
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)