Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons
Posted on: 12 July 2019 , updated on: 15 July 2019
Preprint posted on 17 June 2019
Article now published in eLife at http://dx.doi.org/10.7554/eLife.48718
Coupling of guidance cue receptors with specific mRNA/ribosome complexes assures rapid selective protein synthesis upon cue stimulation in axonal growth cones.
Selected by Andreas van ImpelCategories: developmental biology, neuroscience
Background
A classical hallmark of directional cell migration is the polarization of the migrating cells with a protrusive active migration front and a retracting back of the cells. Extracellular attractive or repulsive signals directly impact the formation and maintenance of this polarity and therefore the direction of migration. In various migrating cell types specific mRNAs have been shown to accumulate in a polarized fashion within the protrusive active front, which enables the cells to locally translate proteins that affect the dynamics of the actin cytoskeleton, for example. In neurons, the main cell body can be considerably further away from the protrusive active growth cone of the axon, hence locally translating proteins controlling the migration machinery assures that the cells can rapidly react on different cues from the environment on the local proteome level [1-3].
How the activation of guidance cue receptors on the cell surface triggers the spatially restricted subcellular translation of these localized mRNAs is not fully understood today. A previous report showed that in commissural axons the Netrin-1 receptor DCC directly associates with ribosomes, suggesting that the link from the cell surface receptor to the translation machinery can be very direct [4]. In how far such a receptor-ribosome coupling represents a more common mechanism that is employed by multiple receptors in different cell types to trigger locally restricted, selective protein synthesis remains elusive.
Key findings
Koppers and colleagues investigated in the current preprint, whether the previously reported association between the DCC receptor and ribosomes is also evident in another system and whether such complexes would also be formed by other guidance receptors that had been implicated to control local axonal protein synthesis. To this end, they performed co-immunoprecipitation experiments for DCC, Nrp1 (the receptor for Sema3A) and Robo-2 (the Slit2 receptor) on Xenopus laevis brain and eye lysates, followed by mass spectrometry to identify the pulled-down interaction partners. Using this approach, the authors find that all three receptors interact with specific ribosomal proteins in a mRNA dependent manner. The EphB2 receptor, however, that does not act via controlling local protein translation, does not associate with ribosomal proteins in this experimental setup, indicating that such interactions may be specific to receptors that convey their signal via controlling the local protein production within the growth cone. Interestingly, the receptors seem to bind to individual sets of RNA-binding proteins, posing the possibility that these specific interactions control which ribosome/mRNA complexes are linked to a particular receptor. In line with this idea, the Holt lab found that DCC and Nrp1 associate with specific sets of mRNAs which at least partially correspond to known target mRNAs of the pulled-down RNA-binding proteins.
To challenge these findings from whole brain lysates on a cellular level, the authors investigated whether these protein interactions are also evident in retinal ganglion cells. Using proximity ligation assays (PLA), they confirm specific interactions between particular ribosomal proteins and DCC or Nrp1, respectively. Importantly, ligand stimulation of the receptors shows an impact on their binding to the ribosomal proteins, as the PLA signals strongly decrease after addition of the respective ligands to the medium, suggesting a rapid release of ribosome/mRNA complexes after receptor activation. An extracellular cue that has been previously shown to negatively impact local protein translation is EphrinA1, which antagonizes the Netrin-1/DCC induced protein synthesis in hippocampal axons. In line with this function, Koppers et al. report that co-stimulation of Netrin-1 with EphrinA1 does not result in a release of the ribosomal proteins from the DCC receptor but suppresses the increase in target protein levels that is detectable after stimulation with Netrin-1 alone. These results therefore indicate that the effect of different or opposing guidance cues can be integrated at the receptor-ribosome dissociation level, thereby controlling the selective translation of receptor specific mRNAs.
Taken together, the here presented findings pose a model (see Fig. 1) in which axonal guidance cue receptors can bind to specific subsets of mRNAs via their interaction with distinct RNA-binding proteins. Upon ligand activation of the receptor, the RNA/ribosome complexes are released resulting in the local translation of the free mRNAs.
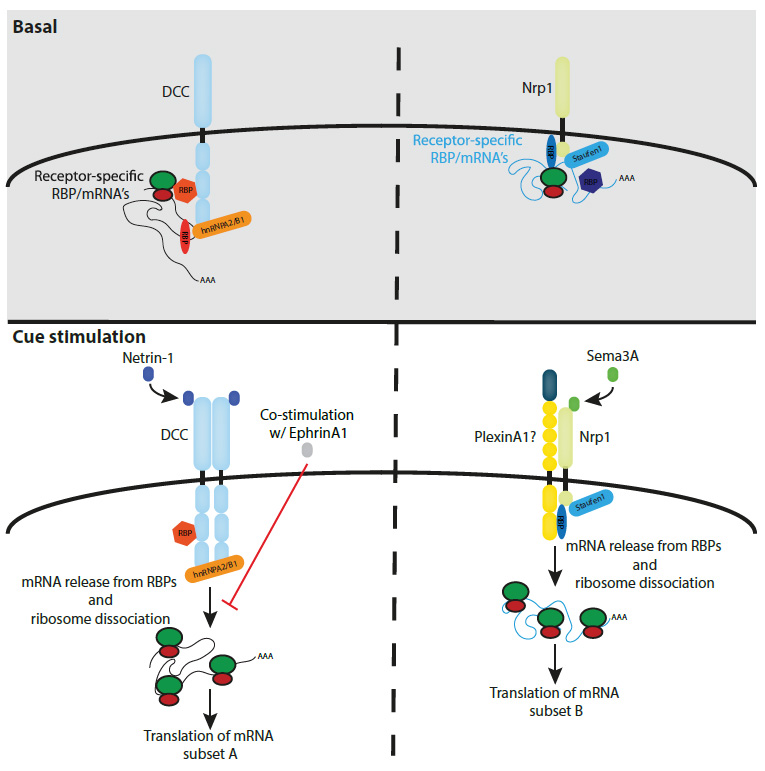
Fig. 1: Axonal guidance cue receptors bind specific sets of mRNAs, ribosomes and RNA-binding proteins, which are released upon cue stimulation leading to the local translation of the mRNAs (reproduced from Koppers et al., Fig. 5B).
Why this is cool
During cell migration attractive and repulsive extracellular signals can not only affect the activity or stability of intracellular proteins but can also influence the local proteome by triggering translation of mRNAs in certain subcellular regions. The results from the Holt lab show that the concept of tethering mRNA/RNA-binding protein/ribosome complexes directly to cell surface receptors is employed by multiple guidance cue receptors in different neuronal cell types and is also conserved in different species. It represents a mechanism by which activation of the receptor is instantly transduced into specific changes of the local protein composition which in turn can influence e.g. the velocity or directionality of the cell movement. Given the finding that subcellular targeting of specific mRNAs is an essential feature in many types of migrating cells including sprouting endothelial cells, for example, the here confirmed model of receptor-ribosome coupling constitutes a concept that might apply to cell movements in a broad variety of cell types and tissues.
Open questions
How is the receptor-ribosome interaction regulated on the molecular level, does e.g. the phosphorylation status of the receptor affect its association with ribosomal proteins?
How do negative signals (EphrinA1) interfere with the receptor-ribosome interaction?
How is a premature translation of the receptor bound form of mRNAs prevented?
Do additional guidance cue receptors also directly bind to ribosomes in other non-neuronal migrating cell types?
Further reading
[1] Liao, G. et al., Control of cell migration through mRNA localization and local translation. WIREs RNA, 6:1–15. (2015)
[2] Cioni, J.M. et al., Molecular control of local translation in axon development and maintenance. Curr Opin Neurobiol. 51:86-94. (2018)
[3] Cagnetta, R. et al., Rapid Cue-Specific Remodeling of the Nascent Axonal Proteome. Neuron, 99, 29-46 (2018)
[4] Tcherkezian, J. et al., Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell, 141, 632-44. (2010)
doi: https://doi.org/10.1242/prelights.12001
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Cooperation between cortical and cytoplasmic forces shapes planar 4-cell stage embryos
Corentin Mollier, Shivani Dharmadhikari
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
Also in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)