A single-cell transcriptomics CRISPR-activation screen identifies new epigenetic regulators of zygotic genome activation
Preprint posted on 21 August 2019 https://www.biorxiv.org/content/10.1101/741371v1
Article now published in Cell Systems at http://dx.doi.org/10.1016/j.cels.2020.06.004
Finding regulators of zygotic genome activation through CRISPR-activation screening
Selected by Sergio MencheroCategories: developmental biology, genomics
Background & Summary:
After the first cell division that takes place in the mouse zygote, when the embryo consists of two cells, a critical event in development occurs: zygotic genome activation (ZGA). This transition coincides with the reprogramming of histone modifications, chromatin accessibility, as well as global DNA demethylation, and results in the transcriptional activation of thousands of genes. Some regulators of ZGA have been identified, but a global understanding of the regulation of this major event is still lacking.
The authors of this preprint use CRISPR-activation (CRISPRa) in a high-throughput screen in mouse embryonic stem cells (ESCs) to identify novel regulators of ZGA. This tool is based on the targeting of a dead Cas9 (dCas9) to gene promoters using short-guide RNAs (sgRNAs) to upregulate gene transcription. The authors targeted 230 candidate regulators of ZGA and after single-cell RNA-sequencing (scRNA-seq) of CRISPRa-perturbed cells, they identified 44 putative regulators that activated a ZGA signature. The authors then selected three of them: Dppa2, a DNA-binding factor already associated to ZGA by the same lab (Eckersley-Maslin et al. 2019), the chromatin remodeller Smarca5 and the top-ranking transcription factor Patz1; and confirmed that their activation resulted in a large fold-change in ZGA-signature genes. After analysing loss of function and rescue experiments using Dppa2 and Smarca5, the authors concluded that the chromatin remodeller Smarca5 regulates ZGA in a Dppa2-dependent manner.
Major experiments in the preprint.
- CRISPR-activation screening of candidate regulators of zygotic genome activation. The authors targeted 230 candidate genes which are expressed prior to and at the time of ZGA based on available datasets of mouse embryos or related to the ZGA-like state of mouse ESCs. Mouse ESCs were transduced with a lentiviral library containing selected sgRNAs and this was followed by scRNA-seq. After multi-omics factor analysis, they identified a set of 44 factors that promoted a ZGA-like response, including the upregulation of known ZGA associated elements such as MERVL or ERV1.
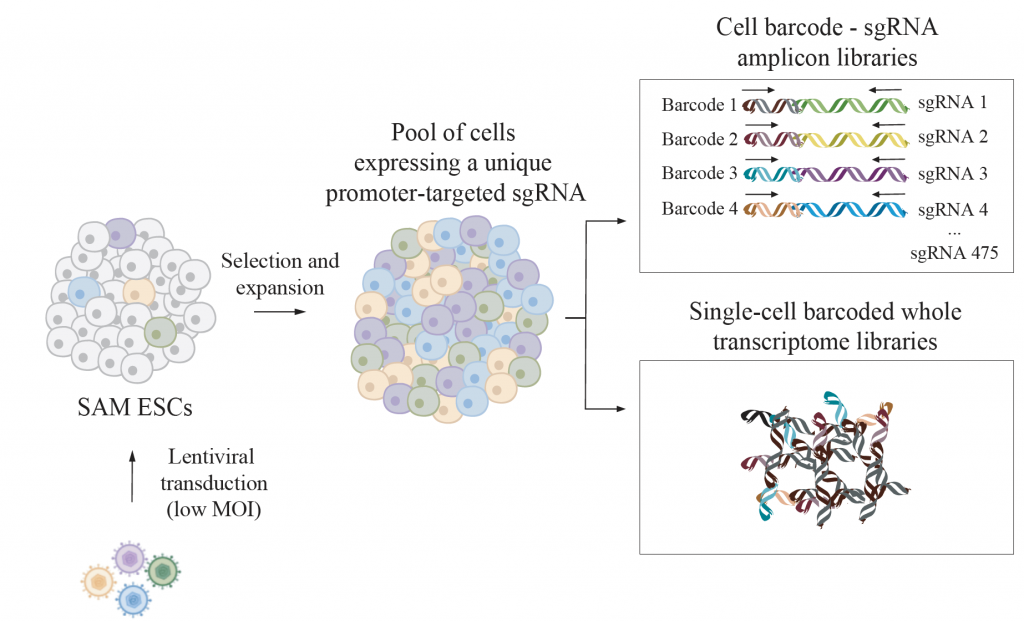
- Validation of candidate regulators of ZGA: Dppa2, Smarca5 and Patz1. The authors induced overexpression of their three candidate regulators either by individual CRISPRa again or by cDNA-eGFP overexpression followed by bulk RNA-seq. Induction of either of the three factors triggered a similar response and included genes highly expressed at the time of ZGA in mouse embryos.
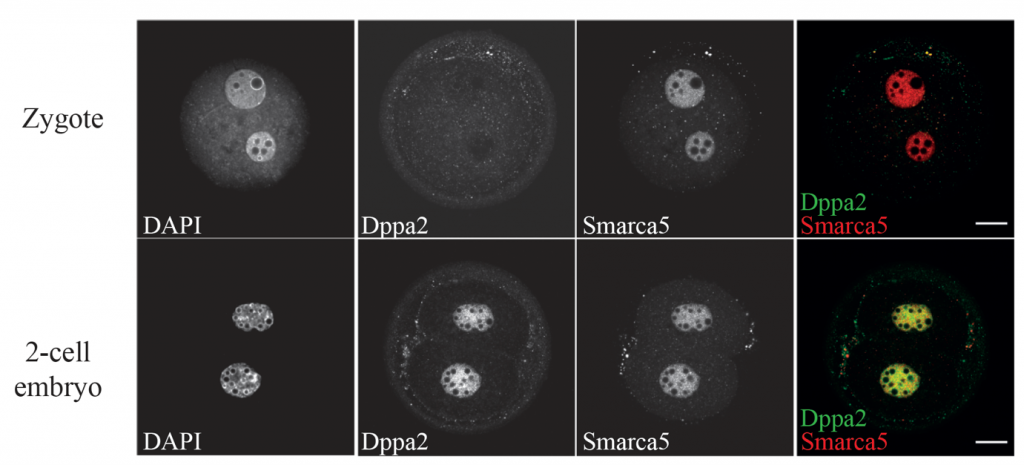
- Smarca5 and Dppa2 are detected in the nuclei of 2-cell stage embryos. Smarca5 is expressed in mouse zygotes in both pronuclei, while Dppa2 is absent in the pronuclei. By the 2-cell stage, Dppa2 starts to be expressed in the nuclei co-localizing with Smarca5.
- Dppa2 is required for Smarca5-mediated regulation of ZGA. Lack of Smarca5 or Dppa2 in ESCs led to a downregulation of ZGA genes. Lack of Smarca5 could be rescued by Dppa2 In contrast, Smarca5 overexpression in Dppa2 mutant ESCs did not rescue the expression of ZGA-related genes
General comments and why I chose this preprint:
The use of a dead-Cas9 to perform CRISPR-activation assays is a very powerful technique and has been shown to activate gene expression at physiological levels (Chavez et al. 2015, Sanson et al. 2018). The authors use this method in a smart way by combining it with high-throughput screening. Also, they overcome the limitations of performing high-throughput screens in 2-cell embryos by successfully using embryonic stem cells. This work exemplifies how there can be major advantages of using a stem cell model system to find interesting candidate regulators for a process happening in embryos. After their initial experiments in ESCs, the authors point out clear candidates that may be studied further in vivo.
This strategy can be applied to multiple contexts to identify regulators of specific processes and complement loss of function perturbations.
Questions to the authors
- The authors use serum/LIF conditions to culture mouse embryonic stem cells (ESCs). Given the different possibilities described in the literature to grow ESCs and achieve a naïve state or a 2C-like state (Macfarlan et al. 2012, Ying et al. 2008), how did the authors decide which was the best option to culture their cells in order to identify regulators of zygotic genome activation? How do they think it could affect the identification of genes in other culture conditions?
- Transcriptional activation of Dppa2, Smarca5 and Patz1 lead to an upregulation of ZGA-related genes. Have the authors found if among the downregulated genes there are known or candidate genes that block ZGA?
References
Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12(4):326-8.
Eckersley-Maslin M, Alda-Catalinas C, Blotenburg M, Kreibich E, Krueger C, Reik W. (2019). Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev 33(3-4):194-208.
Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487(7405):57-63.
Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG. (2018). Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9(1):5416.
Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453(7194):519-23.
Posted on: 11 September 2019
doi: https://doi.org/10.1242/prelights.13748
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Also in the genomics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
preLists in the developmental biology category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genomics category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |











 (No Ratings Yet)
(No Ratings Yet)