EllipTrack: A Global-Local Cell-Tracking Pipeline for 2D Fluorescence Time-Lapse Microscopy
Posted on: 15 June 2020
Preprint posted on 13 April 2020
Article now published in Cell Reports at http://dx.doi.org/10.1016/j.celrep.2020.107984
Categories: cell biology
Background
Biological processes are highly dynamic and display varying degrees of cell-to-cell heterogeneity. Time-lapse microscopy provides the opportunity of tracking and monitoring single-cell dynamics. However, rapid cell migration, high cell density, and treatments or modifications that alter cell morphology and/or behavior make it difficult to track single cells. Multiple tools for cell tracking have been generated. A classical tracker usually consists of three steps: segmentation, in which nuclei (or other structures) are identified; track linking, whereby nuclei (or other structures) are mapped throughout the time-lapse; and signal extraction, whereby fluorescent signals are extracted from regions of interest in each cell to obtain relevant biological quantitative data.
Over the last decade, various efforts have focused on improving the accuracy of each step, however tracking remains challenging due to the high accuracy needed for analysis. Often, manual verification and correction is still required, which limits the experimental throughput. An important breakthrough achieved in cell tracking was the creation of the global track-linking algorithm by Magnusson and colleagues (1). This algorithm uses machine learning tools to infer the probability of cell overlapping, cell migration, and biological events such as mitosis, apoptosis, etc, and then uses the Viterbi algorithm (a dynamic algorithm that aims to compute the most probable path) to iteratively search for, and assemble, the cell track that results in the greatest increase in the probability of existing cell lineages, until this probability can no longer be improved.
However, the use of Viterbi algorithm is a source of tracking mistakes, which Magnusson et al (1) fixed by introducing a swap operation whereby the algorithm examines whether swapping a cell track with another existing one will increase the overall probability of cell lineages. While the algorithm introduced by Magnusson et al increases the accuracy of cell tracking in a small-to-medium scale, it is only partially useful in very high-throughput experiments spanning multiple days, multiple rounds of replication and high cell densities. In the present work, Tian and colleagues (2) address this shortcoming and present EllipTrack, a global-local cell-tracking pipeline optimized for tracking cells in complex time-lapse-derived movies.
Key findings and developments
EllipTrack implements a global track-linking algorithm to construct tracks that maximize the probability of cell lineages, and then corrects tracking mistakes with a local track-correction algorithm (module) whereby tracks generated by the global algorithm are systematically examined and amended if a more probable alternative is found. The new algorithm optimizes the Magnusson swap operation, and offers a higher capacity of correcting tracking mistakes in multi-day, large-scale movies with densely populated cell cultures.
EllipTrack is based on the three-step procedure conventionally used in tracking, but introduces advanced features for each step. For track correction, the novel algorithm presented in EllipTrack consists of multiple steps, each addressing various aspects of tracking mistakes:
The core step optimizes the Magnusson swap operation to fix the mistakes in densely populated regions, using an iterative approach whereby tracking mistakes are progressively fixed.
For corrections, EllipTrack will consider the following alternatives:
- swapping two cell tracks as done by the Magnusson swap operation.
- assigning one cell track for mitosis while terminating the other one.
- swapping two cell tracks and then breaking one of them into pieces.
In addition to the core step, the local track-correction module also fixes tracking mistakes related to under-segmentation, over-segmentation, and undetected mitosis events.
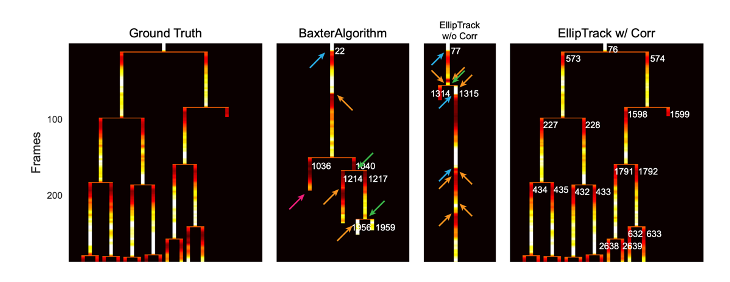
The authors tested the power of EllipTrack by comparing the tracking outcomes with the BaxterAlgorithm. They applied this algorithm to a 48h movie of mammary epithelial cells expressing a nuclear marker and a cell-cycle marker, and showed that EllipTrack could correct tracking mistakes of the BaxterAlgorithm. The authors then examined the ability of the EllipTrack pipeline to identify error-free cell lineages. For this purpose, they measured CDK2 activity as a marker of cell cycle. Using the algorithm, it is possible to evaluate the quality of identified cell lineages by examining whether they match the expected CDK2 activity. They determined that EllipTrack allowed correct identification of cells per lineage.
A systematic benchmarking of EllipTrack against various existing tracking algorithms was performed. These were applied to a variety of cells (HeLa cells, BJ5TA cells, RPE-hTERT cells, MCF10A cells and A375 melanoma cells), under various biological conditions, cell densities, treatments with various drugs, and time-frames, covering multiple generations of cells. The cell trackers were benchmarked on six different criteria:
- SEG: segmentation quality
- TRA: tracking accuracy
- %CORR_S: percentage of cell nuclei correctly segmented before track linking
- #COMP: number of complete tracks where cells were continuously tracked throughout the movie
- #MIS_T: average number of mistakes among the complete tracks
- %CORR_T: percentage of complete tracks correctly tracked
The authors discuss the advantages and shortcomings of each tracking tool, and conclude that EllipTrack is a powerful tool capable of tracking nearly error-free cell lineages. EllipTrack also addresses a shortcoming of various other tools, which assume a constant migration speed of cells over an entire movie. This assumption can result in various tracking mistakes. To account for variable cell behavior, EllipTrack implements an option to perform time- and density-dependent inference of cell migration speeds from training datasets, which users can select for analysis. The authors demonstrate that the time- and density- dependent inference option, in addition to the local track-correction module, allows users to achieve a better tracking performance.
A further advantage of EllipTrack explored by the authors is that it requires minimal training, while still preserving high predictive power, a fact that represents significant time saving for users. Moreover, the authors identified that many tools used for tracking are not always user-friendly and this influences usage and learning. To improve practical usability, the authors developed two user friendly graphical user interfaces for parameter generation and training data.
What I like about this preprint
I like that the authors identified shortcomings associated with cell tracking, and developed their own tool to address those shortcomings. I think they systematically introduce the advantages and uses of EllipTrack in their manuscript. I like that they tested it in multiple biological conditions, and compared it with multiple existing tracking tools. I found the manuscript easy to read, easy to understand, and EllipTrack easy to use. I like also that the authors addressed a big issue affecting use of various image analysis tools: user friendliness. In their work they made sure that EllipTrack was easy to train, and was user friendly. They make their work fully available at github.com/tianchengzhe/EllipTrack.
Open questions
- You tested EllipTracks’ ability to perform in different time- and density-dependent conditions. Is there a limit for either, upon which you find more errors even using EllipTrack?
- You performed a thorough evaluation of EllipTrack in multiple cell lines. This allowed you to see the performance of EllipTrack in cells of different morphologies, migration characteristics, mitotic characteristics, etc. You discuss among the limitations of EllipTrack that cells with kidney shapes or non-elliptical nuclei are over-segmented. Are you planning an improvement on this on future versions, so that users can even choose the cell type being analysed? This would be comparable to the introduction you did of time- and density-dependence.
- In all the cells you used, the nuclei of these cells are of considerable sizes. Is it possible to use EllipTrack to track much smaller cells? And what would the error rate be in such instances? for instance, since you discuss the potential of EllipTrack for drug discovery, I was thinking drug-testing studies in parasitology (including parasites such as Plasmodium or Toxoplasma), although this would involve tracking nuclei under 1µm diameter. Is this possible?
- You mention in the discussion that EllipTrack does not consider unusual cell behaviours and you mention multipolar mitoses and cell-cell fusion as examples. Are there other specific instances where EllipTrack might have performance limitations?
- A huge advantage of your work is that it is open access. Is it possible for users to contribute to the further development of EllipTrack?
- How successful is EllipTrack for tracking fast-moving cells in 3D?
References
- Magnusson, KEG, et , Global linking of cell tracks using the Viterbi algorithm, IEEE Trans Med Imaging, 34, 2015.
- Tian C, et al., EllipTrack: a global-local cell-tracking pipeline for 2D fluorescence time-lapse microscopy, bioRxiv, 2020.
doi: https://doi.org/10.1242/prelights.21943
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)