The collapse of the spindle following ablation in S. pombe is mediated by microtubules and the motor protein dynein
Posted on: 24 June 2021 , updated on: 16 August 2023
Preprint posted on 20 May 2021
Article now published in Biophysical Journal at https://doi.org/10.1016/j.bpj.2021.12.019
Ablating the microtubule spindle during cell division causes it to collapse. Zareiesfandabadi et. al. show how the minus-end directed motor protein dynein, plays a role in this collapse.
Selected by Leeba Ann ChackoCategories: cell biology, microbiology
Background:
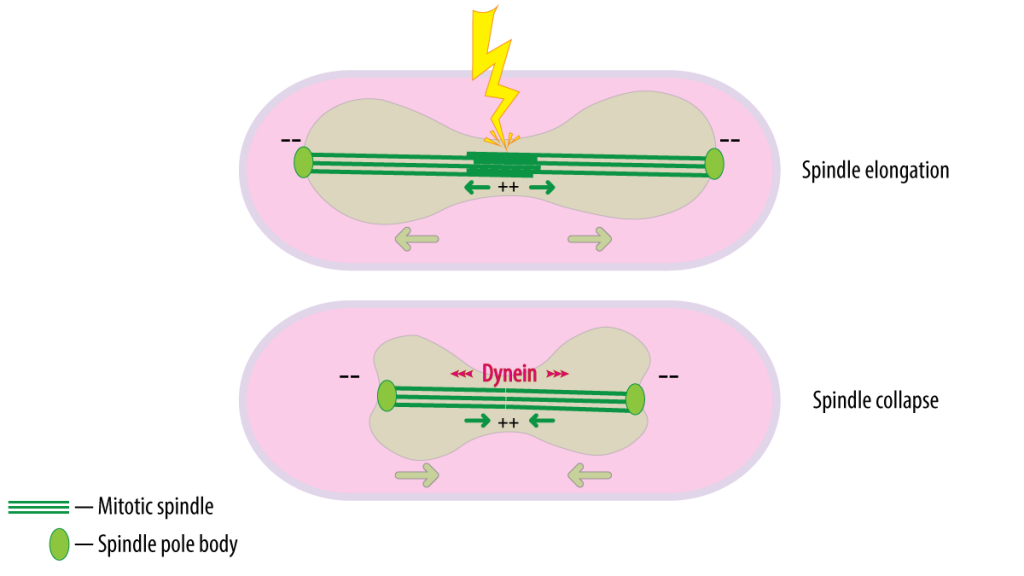
Schematic depicting the collapse of the spindle after laser ablation which is aided partially by dynein.
Unlike many of their mammalian counterparts, fission yeasts are unicellular eukaryotes that undergo ‘closed mitosis’, where the nuclear envelope does not undergo breakdown during cell division. Instead, at the onset of mitosis, cytoplasmic microtubules are reorganized to form the mitotic spindle within the closed nucleus1. This spindle elongates inside the nucleus causing it to expand and in the process, the DNA from the mother cell is equally distributed into identical daughter cells.
Scientists have wondered whether the nucleus and/or its contents can exert an opposing force to the elongating spindle. Previous work has shown that ablating the mitotic spindle with a laser causes the spindle to collapse2,3. This collapse was suggested to occur due to passive viscoelastic relaxation of the nucleus and/or mechanical relaxation of stretched chromosomes. However, in this preprint, Zareiesfandabadi et. al. show that active forces driven by dynein contribute to the collapse of the spindle.
Dynein is known to aid chromosome biorientation to enable its proper segregation in fission yeast4,5. Here, Zareiesfandabadi et. al. demonstrate dynein’s role in ensuring mechanical force balance in the spindle through its ability to slide antiparallel microtubules towards each other as the spindle elongates in the opposite direction.
Key results:
Zareiesfandabadi et. al. used the technique of laser ablation to split the spindle in the middle to identify the forces being exerted on the broken, collapsing spindles.
Upon ablating the spindle, they showed that the collapse observed post-laser ablation follows an exponential relaxation response. While this observation is consistent with viscoelastic relaxation, it did not explain the observed spindle rotation within the nucleus. To test whether active forces are at play, the authors examined how much passive nuclear and chromosomal relaxation forces contribute to spindle collapse.
The authors found that upon ablating the early-stage, short spindle, the separated spindles collapsed towards each other while the chromosomes remained largely stationary. Contrastingly, when ablating the later-stage, long spindle, the chromosomes moved along with the collapsing spindle. Interestingly, the authors observed inward indentations in regions of the chromosome that were connected to the spindle ends. Similarly, there were inward indentations on the surface of the nuclear envelope that was in close proximity to the spindle ends. Based on these results, the authors concluded that neither mechanical relaxation of chromosomes nor viscoelastic relaxation of the nucleus contributed to the collapse of the spindle.
The authors determined that while actin plays no role in spindle collapse, microtubule dynamics are necessary for spindle collapse. They found that the minus end-directed motor protein, dynein, aids spindle collapse. In the absence of dynein, the spindle opts for a prolonged rotational diffusion instead of exhibiting a relaxation response. From this, the authors concluded that the observed collapse of the spindle post-ablation is partially mediated by active forces from dynein and microtubule polymerization.
What I liked about this preprint:
What stood out most for me was the elegance and simplicity of the experiments that were used to test the hypothesis. The authors were able to uncover substantial molecular dynamics using the well-known technique of laser ablation. These experiments reiterate the importance of devoting time towards analyzing data extensively so that one does not miss out on discoveries like the inward indentations the authors observed.
References:
1. Mehta, K., et al., Association of mitochondria with microtubules inhibits mitochondrial fission by precluding assembly of the fission protein Dnm1. J Biol Chem, 2019. 294(10): p. 3385-3396.
2. Khodjakov, A., S. La Terra, and F. Chang, Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol, 2004. 14(15): p. 1330-40.
3. Tolic-Norrelykke, I.M., et al., Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol, 2004. 14(13): p. 1181-6.
4. Grishchuk, E.L., I.S. Spiridonov, and J.R. McIntosh, Mitotic chromosome biorientation in fission yeast is enhanced by dynein and a minus-end-directed, kinesin-like protein. Mol Biol Cell, 2007. 18(6): p. 2216-25.
5. Courtheoux, T., et al., Dynein participates in chromosome segregation in fission yeast. Biol Cell, 2007. 99(11): p. 627-37.
6. Schreiner, S.M., et al., The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun, 2015. 6: p. 7159.
doi: https://doi.org/10.1242/prelights.29767
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Cell cycle-dependent mRNA localization in P-bodies
Mohammed JALLOH
Control of Inflammatory Response by Tissue Microenvironment
Roberto Amadio
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the microbiology category:
Characterization of natural product inhibitors of quorum sensing in Pseudomonas aeruginosa reveals competitive inhibition of RhlR by ortho-vanillin
UofA IMB565 et al.
Feedback loop regulation between viperin and viral hemorrhagic septicemia virus through competing protein degradation pathways
UofA IMB565 et al.
Lytic bacteriophages interact with respiratory epithelial cells and induce the secretion of antiviral and proinflammatory cytokines
UofA IMB565 et al.
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)