Clathrin adaptors mediate two sequential pathways of intra-Golgi recycling
Posted on: 26 March 2021 , updated on: 29 March 2021
Preprint posted on 17 March 2021
Article now published in Journal of Cell Biology at http://dx.doi.org/10.1083/jcb.202103199
Categories: cell biology
Background:
In eukaryotes, proteins are made within the endoplasmic reticulum (ER), an organelle composed of tubules and flattened sacs in the cytoplasm, and then transported to an organelle known as the Golgi Apparatus (GA). Within the GA, proteins can be further processed, for example, they can become glycosylated via the addition of carbohydrate groups by enzymes within the GA. The GA itself is made up of sub-compartments called cisternae which look like stacks of flat, pouch-like structures. Typically, GA cisterna have two ‘faces’: a cis face, close to the ER, and a trans face, near the plasma membrane of the cell. Proteins enter the GA via the cis end, progress through the organelle (during which time the protein is modified) and emerge from the trans-Golgi network (TGN) at the trans end to be journeyed elsewhere (1). In some yeasts, the GA is not stacked, with individual cisterna seen maturing from cis-to-trans (2).
Though the GA is well studied, we still don’t fully understand how cargo transports through this organelle or how recycling of GA transmembrane proteins (needed for the reactions/cargo movement) occurs. Largely, two models exist to explain cargo flow: 1) The Vesicular Transport (VT) model, and 2) the Cisternal Maturation (CM) model. The VT model suggests cargo moves through the GA from the cis to trans faces inside vesicles (a structure in which fluids or gases are encompassed by layers of lipids). As they move through, they are modified appropriately. The CM model suggests that the cisternae themselves mature from cis to trans and the cargo is simply a passenger inside, becoming modified during cisternal maturation as they alter their biochemistry (3). Though a CM model is favoured, it is hard to explain why transmembrane GA proteins remain concentrated within distinct regions i.e at the cis, the medial, or the trans end. The authors think this could be due to how these transmembrane proteins are recycled, suggesting the recycling route is what determines where, along the maturation timeline, these transmembrane proteins are placed.
Key Findings:
Throughout this study, the authors use high resolution confocal microscopy (4D microscopy) to trace, over time, the kinetic dynamics (i.e movements) of Golgi transmembrane factors. They performed these experiments in living yeast (S. cerevisiae). In this species of yeast, the GA is not stacked and the cisternae are dispersed in the cytoplasm, meaning they can monitor GA protein dynamics more clearly (2). They examined how the movement kinetics of transmembrane factors in single cisternae alter during maturation when they disrupt recycling pathways.
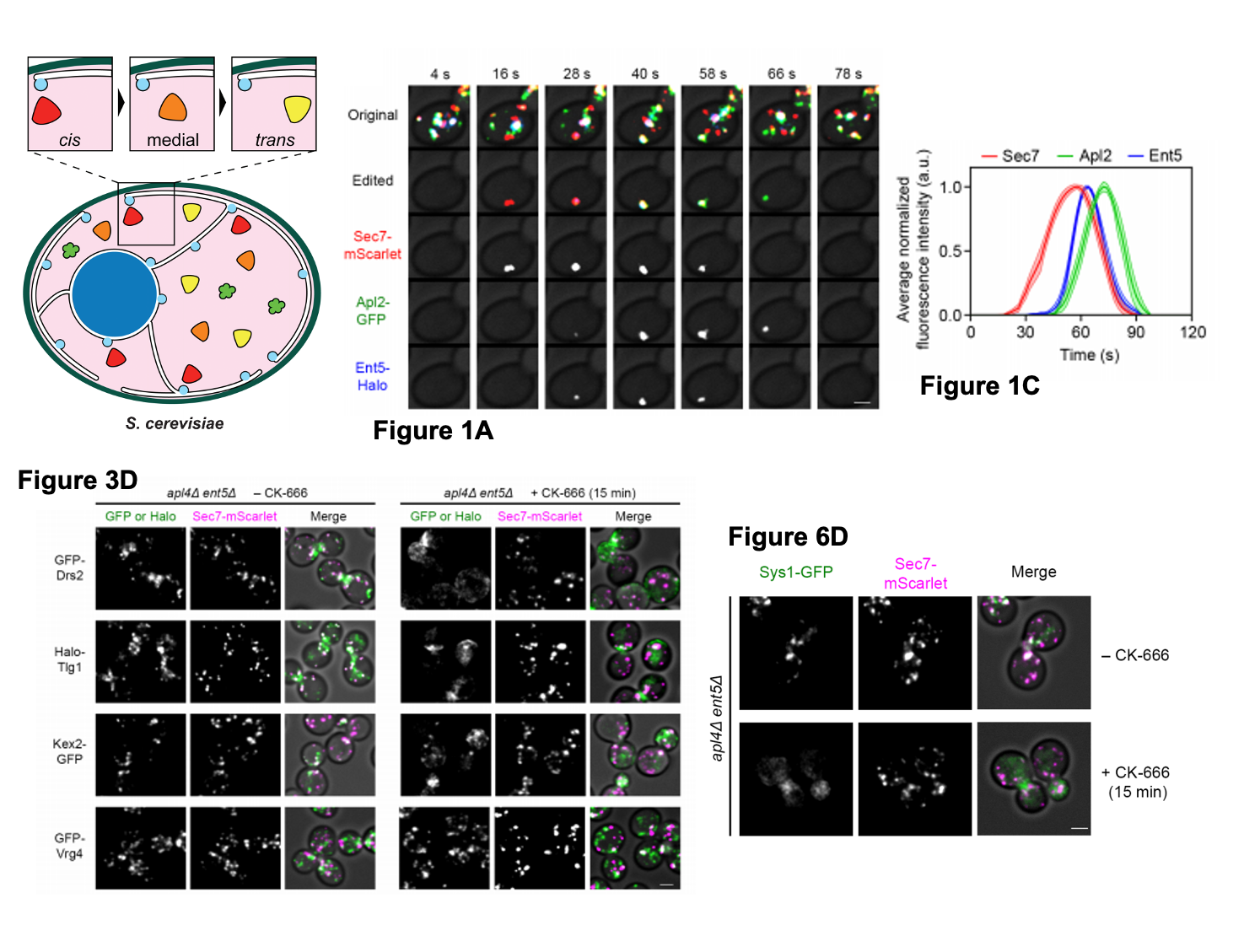
Image shows a selection of data from Casler et al. The schematic representation of the GA arrangement in S. cerevisiae was adapted from Suda and Nakana, 2011 (5). Figure 1A shows the kinetic signatures of AP-1, Ent5 and Sec7 in yeast imaged using 4D confocal microscopy. Figure 1C shows the kinetic signatures of AP-1 (via the subunit Apl2), Sec7 and Ent5 as normalised fluorescent intensity traces. Figure 3D shows representative images detailing the localisation of TGN transmembrane proteins in the absence of AP-1/Ent5 activity and additionally following loss of endocytosis function (by inhibiting with CK-666). Figure 6D shows the mislocalisation of Sys1 in AP-1/Ent5 lacking cells in the presence of CK-666. Yeast schematic made available via a CC-BY 4.0 International license. Figures adapted from Casler et al.
- AP-1/Ent5 can act during TGN membrane recycling
First, the authors show that a recycling pathway co-ordinated by the adaptor complex AP-1 and the associated factor Ent5 localise dynamically to maturing trans-Golgi-like cisternae. The signal of Ent5 both appears before and leaves before AP-1 arrival, revealing AP-1 and Ent5 have separate kinetic patterns which overlap in maturing cisternae. They test if AP-1/Ent5 can recycle the membrane in the trans-Golgi by tracking the movements of a fluorescent membrane dye called FM-4-64, which is trafficked through the GA to the TGN. They show that when AP-1/Ent5 are deleted, much less FM-4-64 was found in the TGN suggesting AP-1/Ent5 can facilitate TGN membrane recycling during cisternal maturation.
- When AP-1/Ent5 are lost, TGN membrane recycles via the plasma membrane
Interestingly, AP-1/Ent5 loss is not lethal meaning recycling also happen via an alternative route. The authors asked if the TGN membrane could be recycled via endocytosis to the plasma membrane. By blocking endocytosis in normal cells and cells lacking AP-1/Ent5 using an inhibitor called CK-666, they saw when endocytosis was blocked in AP-1/Ent5 lacking cells, the cisternae almost completely failed to mature. This suggests that TGN recycling is vital and, in the absence of AP-1/Ent5, the TGN membrane recycles via the plasma membrane.
- A subset of transmembrane proteins recycles via the AP-1/Ent5 pathway
Next, the authors identified other AP-1/Ent5 recycling dependent transmembrane Golgi proteins. Previous data supports the recycling of a phospholipid translocase (Drs2) via AP-1 activity. Drs2 is found in the TGN. Here, they show that when endocytosis is blocked using CK-666 in AP-1/Ent5 deleted cells, Drs2 localised to the plasma membrane instead of to the TGN supporting the recycling of Drs2 via AP-1/Ent5. They found no changes in the localisation of a protein called Vrg4, which is recycled in the early Golgi, suggesting distinct recycling pathways. Further investigation revealed a small cohort of transmembrane TGN proteins are recycled via AP-1/Ent5, showing these proteins largely mirror the movements of Drs2 during cisternal maturation. Thus, the movement and recycling of these transmembrane proteins correlates with the timing of an AP-1/Ent5 driven recycling pathway. Proteins which left before Drs2 arrived were not affected by the loss of AP-1/Ent5 reinforcing the presence of separate recycling pathways
- PVE recycled proteins are recycled independently to the actions of AP1/Ent5
Other Golgi transmembrane proteins traffic to the TGN via prevacuolar endosomes (PVE; i.e late endosomes). This is likely independent of AP-1/Ent5. Recently, the authors have shown that the vacuolar hydrolase receptor (Vsp10) exits the GA at the point of transition when a cisterna becomes TGN-like (4). This suggests Vsp10 would leave before Drs2. However, they saw Vsp10 and Drs2 signals overlapping. They also found another protein, Nhx1, behaved similarly to Vsp10. Together, the localisation of both Vsp10 and Nhx1 to the TGN, even in the absence of AP-1/Ent5 and CK-666 treatment, suggests these proteins recycle independently via a separate recycling pathway operating at a largely similar time to the AP-1/Ent5 recycling pathway in the trans-Golgi/TGN.
- AP-1/Ent5 may facilitate a second GA recycling pathway
Lastly, the authors investigated the transmembrane TGN protein, Sys1. They show that Sys1 and Sec7(a late Golgi marker) behave differently. Sys1 appears and leaves before Drs2 meaning Sys1 traffics independently to Sec7 and Drs2 upstream of trans-Golgi maturation. Unexpectedly, Sys1 recycling still required AP-1/Ent5 activity and in their absence, Sys1 recycled via the plasma membrane like Drs2. They next hypothesised that Sys1 signal, in the absence of AP-1/Ent5, should remain until the cisterna becomes a mature TGNs but this is not what they observed. Instead, Sys1 and Sec7 left at the same time. This is interesting as it suggests AP-1/Ent5 may operate a second recycling pathway in the medial/trans Golgi region. They saw a similar effect with two other transmembrane Golgi proteins known as Aur1 and Rbd2.
What I liked about this preprint:
I really enjoyed reading this preprint. We still lack information on how the GA itself maintains and regulates its transmembrane composition, and though models have been proposed to explain this, no consensus has been reached. Here, the authors built upon their previous observations using high resolution live cell imaging to try and address this knowledge deficit. Their data highlights an intimate connection between the GA and its associated recycling pathways whilst raising the possibility of a currently unknown control pathway underpinning GA activities. Investigating this hypothesis further could shed much needed light on the regulation of GA maturation, particularly as some diseased states in humans are associated with GA and trafficking defects.
Questions for the authors:
Q1: You suggest a logic circuit operates to control GA recycling pathways by, for example, turning recycling pathways on and off at appropriate times and you show multiple recycling pathways plays roles in determining what transmembrane proteins arrive at the GA and at what maturation stage. What do you think could be the signal(s) to activate these recycling pathways?
Q2: Is there a defining factor/structure/motif etc that makes a trans Golgi transmembrane protein different from a cis transmembrane Golgi protein? I.e how does the recycling machinery make the decision as to what is going where if the proteins are trafficked to, say the trans-Golgi, independent to the function they perform in the trans-Golgi itself?
Q3: The Golgi is a dynamic organelle. How does the Golgi form its compartments and how are they maintained as they transition from cis-trans?
References:
- H.Mollenhaur and D.J.Morre. Perspectives on Golgi apparatus form and function. J. Electron Microsc. Tech (1991).
- Suda and A.Nakano. The Yeast Golgi Apparatus. Traffic (2011).
- S.Glick and A.Luini. Models for Golgi Traffic: A Critical Assessment. Cold Sping Harbor Perspectives in Biology (2011).
- C.Casler and BS.Glick. A microscopy-based kinetic analysis of yeast vacuolar protein sorting. eLife (2020).
- Y.Suda and A.Nakano. The yeast Golgi apparatus. Traffic. 2012.
doi: https://doi.org/10.1242/prelights.27909
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)