Mutations in Bcl9 and Pygo genes cause congenital heart defects by tissue-specific perturbation of Wnt/β-catenin signaling
Posted on: 7 February 2018 , updated on: 21 February 2018
Preprint posted on 17 January 2018
Article now published in Genes & Development at http://dx.doi.org/10.1101/gad.315531.118
Nuclear regulators of the Wnt/ß-catenin pathway – previously associated with human developmental cardiac malformations - as key players during vertebrate heart patterning.
Selected by Andreas van ImpelCategories: developmental biology, genetics
Background
Genetic alterations in BCL9 and BCL9-like have previously been associated with the development of congenital heart disease in humans, a condition that nearly affects 1% of the population and presents with various structural and functional abnormalities of the heart. BCL9 proteins and their interaction partners PYGO1/2 have been shown to act as co-factors of nuclear ß-catenin in the context of canonical Wnt signalling. Despite the involvement of the Wnt pathway in several aspects of cardiac development, a function for BCL9/PYGO genes during vertebrate heart formation has not been reported so far.
Key findings
Here, the authors of this preprint tackle the question whether BCL9 and PYGO are required for heart development, making use of two vertebrate model systems: zebrafish and mouse. In both species, loss of either protein function results in severe cardiac defects. Importantly, interference with the BCL9/PYGO complex seems to cause a selective reduction of canonical Wnt signalling levels in the heart and other affected tissues but does not systemically abrogate transcriptional activity of ß-catenin suggesting that both proteins are involved in mediating only a specific subset of Wnt signalling events in mouse and zebrafish embryos. Using conditional knock-outs in mice the authors further demonstrate a specific requirement for Bcl9/Pygo in early mesodermal cardiac progenitors and in migrating cardiac neural crest cells, two key lineages during heart development. Mechanistically, the ß-catenin/BCL9/PYGO complex seems to control the expression of a specific subgroup of Wnt target genes including key transcription factors that are essential for different steps during heart patterning.
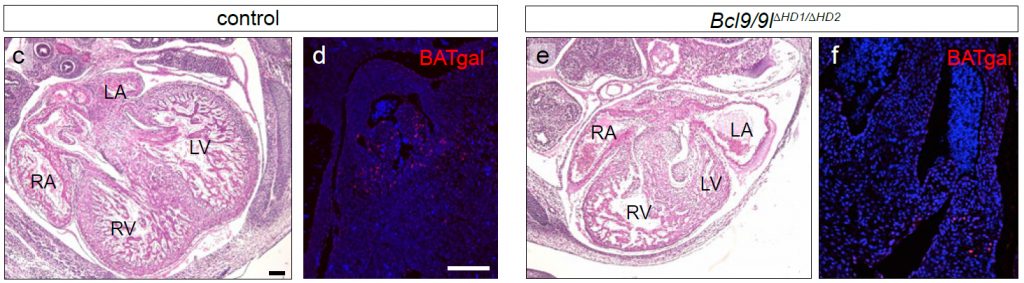
Loss of functional BCL9 proteins in mice results in pronounced heart defects (thinned myocardium of the ventricular walls; malformations of the forming septum, the atrio-ventricular valves and the outflow tract) and a reduced expression of a Wnt reporter (BATgal) in the outflow tract. Reproduced from Cantu et al Fig 3c-f.
How I believe this moves the field forward
The preprint from Cantù and colleagues identifies BCL9 and PYGO as essential tissue-specific co-factors of ß-catenin that selectively mediate the activation of canonical Wnt target genes during vertebrate cardiac development. In my view this is a significant finding as the presented results strongly imply that the reported genomic alterations in BCL9 genes in humans with congenital heart disease are indeed causative or at least contribute to the respective cardiac malformations found in patients. Therefore, this study gives important new insights into the molecular basis of congenital heart disease in humans.
Open questions
- How is the tissue-specific activity of BCL9/PYGO brought about and how is it regulated? Is the (overlapping) expression of both genes restricted to the affected tissues?
- Is there a common set of cardiac target genes downstream of BCL9/PYGO/ß-catenin signalling in mice and zebrafish that might also be misregulated in human patients?
- Is the cardiac function of BCL9/PYGO restricted to early developmental processes or is this complex also required at later stages, e.g. in response to cardiac injury?
Further reading – useful reviews
Grant, M. G., Patterson, V. L., Grimes, D. T., Burdine, R. D. (2017) Modeling Syndromic Congenital Heart Defects in Zebrafish. Curr Top Dev Biol.;124:1-40.
Andersen, T. A., Troelsen, K. de L. L. & Larsen, L. A. (2013) Of mice and men: molecular genetics of congenital heart disease. Cell Mol Life Sci 71, 1327–52.
Mosimann, C., Hausmann, G., Basler, K. (2009) Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nature Reviews. Molecular Cell Biology, 10(4):276-286.
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Gestational exposure to high heat-humidity conditions impairs mouse embryonic development
Girish Kale, preLights peer support
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the genetics category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Lipid-Based Transfection of Zebrafish Embryos: A Robust Protocol for Nucleic Acid Delivery
Roberto Rodríguez-Morales
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
María Mariner-Faulí
preLists in the developmental biology category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genetics category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |











 (No Ratings Yet)
(No Ratings Yet)