Species-specific oscillation periods of human and mouse segmentation clocks are due to cell autonomous differences in biochemical reaction parameters
Posted on: 18 June 2019
Preprint posted on 26 May 2019
Article now published in Science at http://dx.doi.org/10.1126/science.aba7668
Computational Modelling in the era of quantitative developmental biology
Selected by Irepan Salvador-MartinezCategories: bioinformatics, developmental biology, genetics
Introduction
Complex systems are especially well suited to be studied by computer models as their behaviour depends on many variables (or parameters) which interact in a non-linear manner. What this means is that minor differences in a single variable can produce drastic effects on the system’s behaviour. At their best, computer models can produce accurate predictions on the behaviour of such systems given a set of initial conditions. Every day we rely on computer models: when we open our weather forecasting app and decide we should take the raincoat, we are trusting the results of heavily intensive computer simulations that predicted that it will rain today.
Developmental biology is a field where the use of computational modelling has become increasingly popular in recent years. This is totally justified, as developmental processes are perfect examples of complex non-linear systems: their outputs depend on multiple parameters at different levels (biochemical interactions, gene regulatory networks, cell-cell interactions, cell behaviours, physical constraints, etc) that interact in a non-linear manner and that feed back into each other. Multiple types of models are used in developmental biology, but they all share the same purpose: “to treat mechanistic ideas in a more rigorous manner than we are capable with our own minds” (Sharpe, 2017). One type of computational model is the one that allows for hypothesis testing against quantitative data (Sharpe, 2017). In this case, the researcher has a specific hypothesis from the beginning that he/she wants to test. What is needed are quantitative data on the parameters that he/she considers relevant to the model.
About the preprint
In the preprint Matsuda et al. use such a modelling approach that allows hypothesis testing against quantitative data. In their case the hypothesis put to test is the following: Can differences in the speed of biochemical reactions between mouse and human cells explain the timing difference of their segmentation clocks? In vertebrates, the segmentation clock refers to the oscillatory gene expression that regulates the timing of sequential formation of body segments (somites).
To answer this, Matsuda et al. used cultures of pre-somitic mesodermal cells (PSM) of both mouse and human, making sure that the oscillation periods (i.e., the duration of one cycle) of each species were recapitulated in vitro. Using the temporal expression pattern of the gene Hes7 to quantify the oscillation period (Fig. 1), they showed that it was ~2hrs and ~5hrs in mouse and human, recapitulating the 2-3 fold period difference observed in vivo.
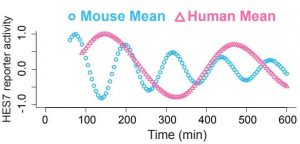
After first demonstrating that the timing differences are cell-autonomous and that these do not depend on the sequence differences between the orthologous genes (see Teresa’s preLight) they focused on the intracellular network that drives gene expression oscillation (Fig. 2). This intracellular network consists of an auto-inhibitory feedback loop of HES7. This means that when HES7 is expressed, it inhibits its own expression (going from ON to OFF); when HES7 is not expressed any more, there is no inhibition so expression starts again (from OFF to ON).
Matsuda et al. used a previously proposed mathematical model (Lewis, 2003) that theoretically showed that auto-repression of a gene by its own product (like HES7) can generate oscillations, with its period determined by transcriptional and translational delays, and degradation rates. For an experimental validation of this model, they carefully quantified the relevant biochemical parameters: the degradation rate of HES7 protein, the transcription and translation delay of HES7, the delay produced by intron processing, etc.
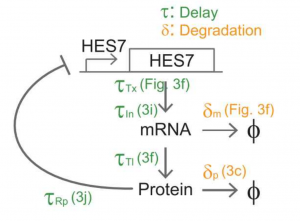
They fed these experimentally determined biochemical parameters into the computational model, generating in this way a mouse and a human version of it that could be directly compared. The simulations showed that the mouse and human in silico periods were ~150min and ~300min, reproducing the observed 2-3 fold period difference in their in vitro experiments. This finally proved that the slower biochemical reactions of HES7 in the human PSM can indeed explain its longer oscillation period, as compared to the mouse.
Why I chose this preprint:
I liked very much this preprint because it is a remarkable example of model testing against quantitative data. The authors carefully quantified the parameters needed to build a more realistic segmentation clock model and were able to prove a hypothesis that has been previously made based on theoretical computational modelling. In doing that, Matsuda et al. built a bridge between theoretical modelling and quantitative developmental biology. Let’s hope we see more studies like this.
References:
Lewis, J. (2003). Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Current Biology, 13(16), 1398–1408. https://doi.org/10.1016/S0960-9822(03)00534-7
Sharpe, J. (2017). Computer modeling in developmental biology: growing today, essential tomorrow. Development (Cambridge, England), 144(23), 4214–4225. https://doi.org/10.1242/dev.151274
Irepan and Teresa’s questions to the authors:
Q1: Since the size and number of somites differ between mouse and human, it would be interesting to know if the identified temporal mechanisms play a role in counting the number or measuring the size of somites. Do the authors think that the in vitro protocol in mouse and human generates the appropriate number of somites? Does the oscillation period vary over time in vitro?
Q2: For the comparative analysis between species, reporter activity has to be normalized. I wonder if the amplitude of Hes7 is different between mouse and human, or if the levels of expression are compensated between species.
Q3: Since the differences in biochemical reaction parameters determine the tempo of the species, do the authors think that each parameter is independent?
Q4: Since the time of development is species-specific, do the authors think that this has any evolutionary advantage for the species?
Q5: Have you considered perturbation analyses (e.g. introduce large introns to increase intron delay or alteration of mRNA turnover rate) as a further validation of the model?
doi: https://doi.org/10.1242/prelights.11311
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Expressive modeling and fast simulation for dynamic compartments
Benjamin Dominik Maier
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
Julio Molina Pineda
Also in the developmental biology category:
Gestational exposure to high heat-humidity conditions impairs mouse embryonic development
Girish Kale, preLights peer support
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the genetics category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Lipid-Based Transfection of Zebrafish Embryos: A Robust Protocol for Nucleic Acid Delivery
Roberto Rodríguez-Morales
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
María Mariner-Faulí
preLists in the bioinformatics category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the developmental biology category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genetics category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |











 (No Ratings Yet)
(No Ratings Yet)