The ELT-3 GATA factor specifies endoderm in Caenorhabditis angaria in an ancestral gene network
Posted on: 11 June 2022 , updated on: 19 December 2022
Preprint posted on 26 May 2022
Article now published in Development at http://dx.doi.org/10.1242/dev.200984
How many ways to make a worm gut? Extensive rewiring of the gene network underlying gut development in Caenorhabditis
Selected by Chee Kiang EweCategories: developmental biology, evolutionary biology, genetics
Updated 28 October 2022 with a postLight by Swathi Arur
When I received and read this manuscript as an editor, I thought this manuscript may be a good fit for Development and sent it out for peer review since I was struck by what seemed like an example of evolution of a gene regulatory network that led to endoderm specification in two distant yet related species (in this case of the nematode Caenorhabhditis). Using a combination of gene expression reporters, single molecule FISH analysis, and either genetic mutants or RNA interference to deplete gene function, the authors argued that the gene regulatory network that led to specification of the endoderm was not the same between C. elegans and C. angaria. In fact C. angaria lacked some ‘critical’ gut specification genes, ‘critical’ as defined by the model nematode C. elegans, which led the group to identify a more basic gene regulatory network which drove endoderm specification in nematodes.
Updated 26 October 2022 with a postLight by Chee Kiang Ewe
This preprint was recently published in Development. In the peer-reviewed article, the authors included an updated high quality whole genome sequence of C. angaria, which will aid future comparative studies with C. elegans. The authors also expanded their smFISH analysis to C. monodelphis and showed that elt-3 is expressed in the embryonic endoderm, as observed in C. portoensis and C. angaria, further strengthening the conclusion that ELT-3 plays a conserved role in endoderm specification in nematodes outside of the Elegans supergroup. Moreover, the authors performed additional genetic analyses and could show that Can-ELT-3 (driven under the end-3 promotor) may rescue endoderm defects in the C. elegans end-1 end-3 double mutant. Strikingly, the authors were able to move C. angaria network into C. elegans by knocking out the entire GATA cascade in C. elegans and replacing it with Can-ELT-3 and Can-ELT-2, which strongly supports the conclusion that Can-ELT-3 and Can-ELT-2 drive endoderm specification and differentiation, respectively. Interestingly, the rescue appeared to be temperature sensitive as it occurred at lower efficiency at 20oC compared to 25oC, suggesting reduced robustness in nematodes with a simple network as was also pointed out in the preLights Q&A. Overall, I think that the published, peer-reviewed paper has significantly improved compared to the preprint. Congratulations to Broitman-Maduro et al.!
Background:
Development is driven by gene regulatory networks (GRNs) that are composed of complex interactions between many transcription factors. The nematode C. elegans has been a great system to study the principals underlying developmental GRN owing to its defined cell lineage and the extensive genetic tools available for manipulating the GRN. Furthermore, comparative studies involving different nematode species spanning the Caenorhabditis genus (Figure 1) have provided insights into the evolutionary variation in developmental mechanisms, and how GRNs might be modified during evolution (Ewe et al., 2020).
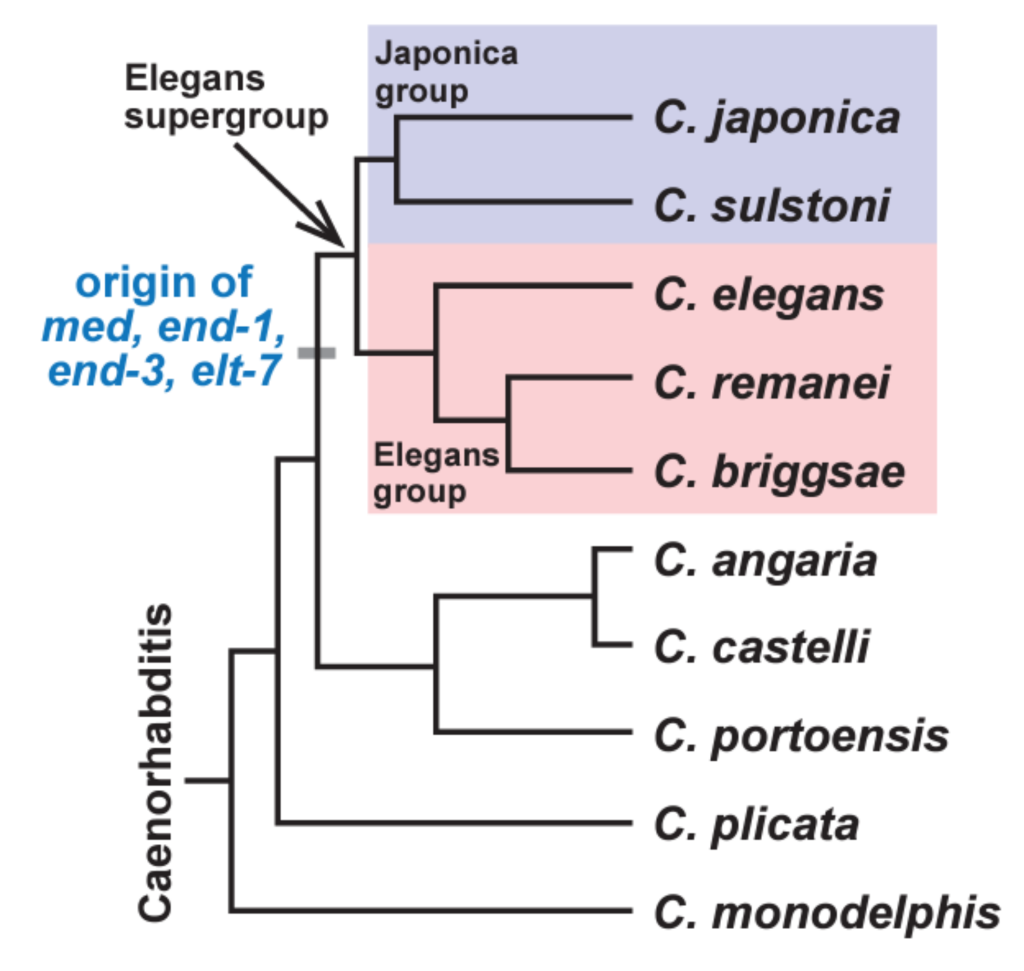
Figure 1: The phylogenetic relationships of the Caenorhabditis nematodes. MEDs, ENDs, and ELT-7 originate at the base of the Elegans Supergroup (adapted from Figure 1; Broitman-Maduro & Maduro, 2022).
In C. elegans, the endoderm comprises the mid-gut. In the early embryo, the endoderm GRN is initiated by SKN-1, which activates a gene cascade driven by six GATA transcription factors – two MEDs, two ENDs, and two ELTs. (Figure 2). Additionally, the Wnt effector POP-1 activates endoderm specification by acting on END-1 and -3. Interestingly, the orthologues of the MEDs and ENDs, as well as ELT-7, are missing in nematodes outside of the Elegans Supergroup, including C. angaria and C. portoensis (Figure 1) (Maduro, 2020), raising the question of how gut is specified in their absence.
In this preprint, the authors found that ELT-3, a GATA factor that is mostly expressed in the hypodermal cells in C. elegans, is present in early endodermal progenitors and drives gut specification in C. angaria. This study demonstrates an extensive rewiring of the endoderm GRN within the Caenorhabditis genus (Figure 2).
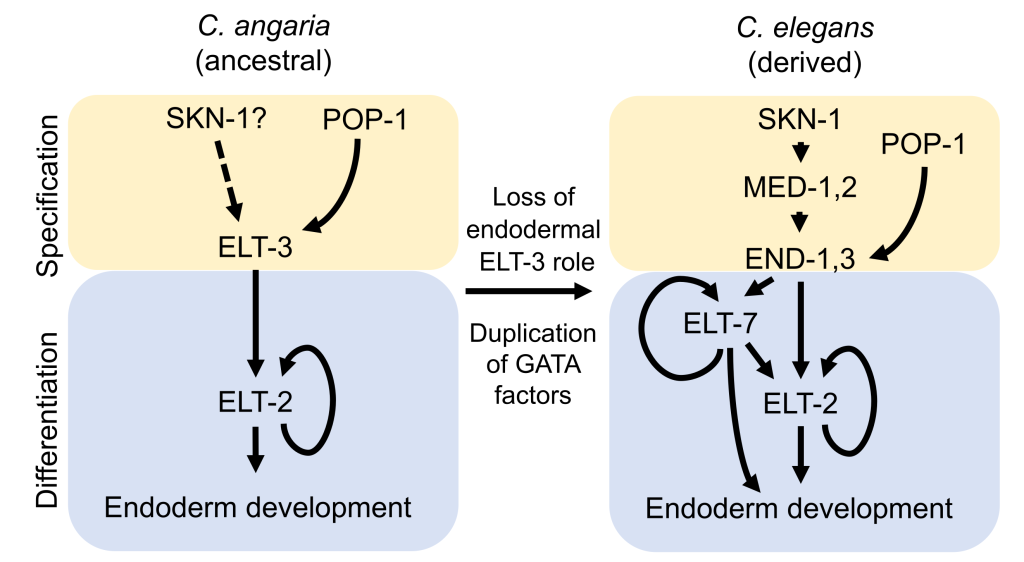
Figure 2: The evolution of the endoderm GRN in nematodes. POP-1 activates ELT-3, which in turns activates ELT-2 in C. angaria. In the derived GRN in C. elegans, MEDs and ENDs, instead of ELT-3, drive endoderm specification. This eventually leads to the activation of ELT-7 and -2, which lock down the differentiated endodermal cell fate.
Major findings:
1) Wnt effector POP-1 is required for embryonic gut development in C. angaria
To test the roles of SKN-1 and POP-1 orthologues in endoderm development in C. angaria, the authors knocked down Can-skn-1 and Can-pop-1 using RNAi. While knocking down Can-skn-1 did not cause obvious developmental defects, embryos lacking Can-pop-1 underwent developmental arrest and failed to development a gut.
2) GATA factor ELT-2 plays conserved role in endoderm differentiation
To examine the role of C. angaria ELT-2 in endoderm development, the authors cloned Can-elt-2 and introduced the transgene into C. elegans. They found that Can-elt-2, like Cel-elt-2, was expressed exclusively in the endodermal lineage. In C. elegans, eliminating both elt-2 and elt-7 causes larval arrest and a severe block to endoderm differentiation (Sommermann et al., 2010). The authors showed that the expression of Can-elt-2 transgene strongly rescued larval lethality in C. elegans double mutant lacking elt-2 and elt-7. Importantly, knocking down Can-elt-2 in C. angaria caused penetrant larval lethality and gut differentiation defects. Together, these results demonstrate the functional conservation of ELT-2 in endoderm differentiation.
3) ELT-3 drives endoderm specification in C. angaria
The authors showed that the expression of Can-elt-2 in C. elegans required END-1 and -3, the two endoderm-specifying GATA factors, as knocking out end-1/3 abrogated Can-elt-2 transgene expression; however, C. angaria does not contain END-1 and -3 orthologues. To identify the upstream activator(s) of Can-elt-2, the authors performed smFISH to examine the expression of other GATA-encoding genes (elt-1, elt-3, and elt-5) in C. angaria embryos. Interestingly, the authors found that the transcripts of elt-3, but not elt-1 and -5, were found in the early endodermal lineage, and the expression of Can-elt-3 depended on Can-POP-1, but not Can-SKN-1.
To examine the role of elt-3 in endoderm specification, the authors overexpressed Can-elt-3 (under the control of a heat-shock promotor) in C. elegans and found a widespread activation Can-elt-2 and ectopic gut development in the arrested embryos. Furthermore, knocked down elt-3 in C. angaria by RNAi resulted in development arrest and a severe block to endoderm development, showing the function of Can-ELT-3 as a potent driver of endoderm specification, akin to END-1 and -3 in C. elegans.
4) The evolution of the endoderm gene regulatory network in nematodes
Lastly, the authors found elt-3 and elt-2 to be expressed in the endodermal lineage of C. portoensis, another species outside of the Elegans Supergroup, suggesting that ELT-3 -> ELT-2 interaction is conserved in the ancestral endoderm GRN, prior to the gene duplication events that gave rise to meds, ends, and elt-7 (Figure 2). The intercalation of the redundant GATA factors in the endoderm GRN may ensure developmental robustness during rapid embryogenesis in C. elegans.
What I liked about this preprint: This work provides an important insight into the dynamics of developmental system drift in the endoderm GRN in nematodes – an excellent paradigm to study the molecular mechanisms of developmental robustness and evolutionary plasticity.
Question for the authors:
The genetic redundancy in C. elegans may indeed promote developmental robustness; the simplicity of the ancestral gene network may therefore result in increased transcriptional noise. Have you noted obvious variability in elt-2 onset timing and/or expression level in C. angaria?
References:
Ewe, C. K., Torres Cleuren, Y. N., & Rothman, J. H. (2020). Evolution and developmental system drift in the endoderm gene regulatory network of Caenorhabditis and other nematodes. Frontiers in Cell and Developmental Biology, 8, 170. https://doi.org/10.3389/FCELL.2020.00170
Maduro, M. F. (2020). Evolutionary dynamics of the Skn-1/Med/end-1,3 regulatory gene cascade in Caenorhabditis endoderm specification. G3: Genes, Genomes, Genetics, 10(1), 333–356. https://doi.org/10.1534/g3.119.400724
Sommermann, E. M., Strohmaier, K. R., Maduro, M. F., & Rothman, J. H. (2010). Endoderm development in Caenorhabditis elegans: The synergistic action of ELT-2 and -7 mediates the specification→differentiation transition. Developmental Biology, 347(1), 154–166. https://doi.org/10.1016/j.ydbio.2010.08.020
doi: https://doi.org/10.1242/prelights.32270
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Gestational exposure to high heat-humidity conditions impairs mouse embryonic development
Girish Kale, preLights peer support
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the evolutionary biology category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Fetal brain response to maternal inflammation requires microglia
Manuel Lessi
Also in the genetics category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Lipid-Based Transfection of Zebrafish Embryos: A Robust Protocol for Nucleic Acid Delivery
Roberto Rodríguez-Morales
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
María Mariner-Faulí
preLists in the developmental biology category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the evolutionary biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the genetics category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)
1 year
Katherine Brown
This paper was recently selected by the editors of Development as a finalist of our 2022 Outstanding Paper prize (see this recent editorial for more details: https://journals.biologists.com/dev/article/150/7/dev201810/306247/)
It’s been great seeing the journey of this paper from preprint to published paper, and to read the comments from Chee Kiang and Swathi on why they both found it an exciting piece of research!