Constricted migration contributes to persistent 3D genome structure changes associated with an invasive phenotype in melanoma cells
Posted on: 2 April 2020
Preprint posted on 26 November 2019
Article now published in EMBO reports at http://dx.doi.org/10.15252/embr.202052149
The costs of breaking and entering: extensive alterations to cells during constricted migration.
Selected by Mariana De NizCategories: cancer biology, cell biology
Background
To move within a complex network made up of tissues and vasculature, migrating cells must squeeze through constrictions of the extracellular matrix or endothelial lining that might be much smaller than their nucleus, requiring major nuclear deformations. Although the cell membrane and cytoplasm are relatively elastic, the ability of a cell’s nucleus to withstand deformations and migrate through small spaces is limited by its size and stiffness. In their preprint, Golloshi et al explored cell migration in the context of cancer and metastasis. Previous evidence suggests that chromosome structure may influence a cancer cell’s ability to undergo confined migration, that changes in chromosome structure could be caused by nuclear deformation, and that chromosome structural changes could influence gene regulation and cancer cell phenotype.
Chromosome structure affects nucleus stiffness, gene regulation and DNA repair. Moreover, nuclear stiffness is affected by lamin and chromatin proteins as well as extracellular forces:
- a) Lamin A/C levels have dual effects depending on the type of cancer: For some cancers, decreased expression of Lamin A/C leads to decreased nuclear stiffness, increased deformability, and higher levels of constricted migration. In other specific cancers, the opposite occurs.
- b) Chromatin has been shown to influence nuclear physical properties, and to play a role in mechano-sensing, nuclear stiffness, and invasiveness. Specifically, increased levels of heterochromatin (firmly packed, genetically inactive form) increase nuclear stiffness while increased euchromatin (uncoiled, genetically active form) levels decrease stiffness.
- c) Extracellular physical forces have also been shown to have significant effects on nuclear deformation.
While several aspects related to cell migration have been well studied, it is yet not fully understood whether the 3D organization of the genome affects or is affected by confined migration, which is the knowledge gap the authors aim to address in this work.
Key findings and developments
Overall findings
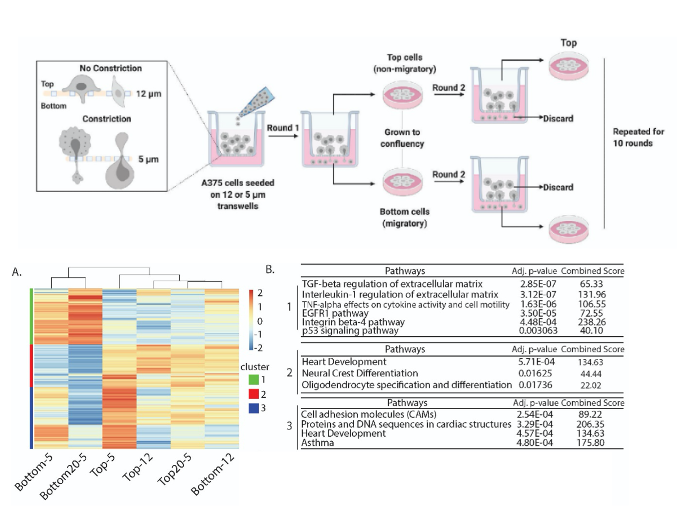
- The authors used invasive human melanoma cells (A375) to investigate the properties of the nucleus and 3D chromosome structure accompanying confined migration. They identified significant phenotypic differences between cells undergoing multiple rounds of constricted migration, compared with those that did not undergo migration through constrictions.
- Cells undergoing constricted migration displayed an abnormal distribution of Lamin A/C and heterochromatin, and had a stable increase in migration efficiency, as well as a specific morphology. These cells also showed changes in chromosome spatial compartmentalization and displayed alterations in expression of genes associated with migration and metastasis.
- Cells that underwent constricted migration also show increased nuclear deformations when cultured in a 3D collagen matrix, and altered behaviour when co-cultured with fibroblasts in organoids.
- Altogether, the study reveals a relationship between chromosome structure changes, metastatic gene signatures, and the abnormal nuclear appearance of aggressive melanoma.
- The study suggests that 3D genome architecture influences the ability of cancer cells to undergo constricted migration and raises the possibility that changes take place after constricted migration which confer a higher metastatic efficiency for subsequent rounds of migration through constrictions.
Specific findings – (if you want to know more…)
Heterogeneous constricted migratory capacity
- The authors performed serial rounds of constricted migration, and found that melanoma cells that successfully undergo constricted migration initially, migrate more efficiently in subsequent rounds.
- The differences in migration efficiency correspond to other notable changes in cell phenotype, including a decrease in cell-cell adhesions, an elongated cell body, and lobopodian/lamellipodia-like migration.
- The authors described in the initial cell population, a fraction of cells that cannot migrate even through a large pore where nuclear deformation would not be required. These cells were described as having higher cell-cell adhesion, and more epithelial-like phenotypes than those cells that did exhibit migratory behaviour.
Differential distribution of Lamin A/C and heterochromatin in cells capable of constricted migration
- It has been reported that, during cancer cell constricted migration, the nuclear envelope can rupture, blebs can form, and DNA damage can occur.
- The authors looked at nucleus structure, and found that in cells capable of constricted migration, the nuclei displayed an altered distribution of Lamin A/C with specific regions of the nuclear envelope devoid of it. They also found that heterochromatin foci in the nuclei of cells capable of constricted migration appear to be more dispersed and the majority of signal is localized to the nuclear periphery
Genomic regions change their compartmentalization after rounds of constricted migration
- To measure changes in genome wide chromosomal contacts, the authors performed genome-wide chromosome conformation capture (Hi-C) on cells with various migratory capacities.
- They observed regions of the genome that switch their compartment identity specifically in the cells capable of constricted migration, but not in any of the other conditions, implying that 3D genome structure changes could be specific to constricted migration.
- Genome-wide hierarchical clustering analysis of the compartment eigenvectors segregates conditions according to whether they have undergone constricted migration or not, indicating that the changes in chromatin compartmentalization are directly linked to migratory ability.
- Gene ontology enrichment analysis of genes in compartment-altered regions identified functions correlating to cell motility and the regulation of actin cytoskeleton.
Gene expression changes after constricted migration reflect metastatic potential
- RNA-Seq was performed on all A375 subpopulations. The authors identified around 1000 genes specifically expressed in cells capable of undergoing constricted migration.
- Upregulated pathways in these cells were related to metastasis.
- Further analyses suggest that a substantial portion of structural changes after constricted migration might be explained by something other than gene regulation changes. The authors hypothesized this might be related to adaptation to physical strain on the chromatin fibre.
Inter-compartment interactions increase after constricted migration
- The relative strength of inter and intra-compartment contacts is different in cells undergoing constricted migration. This change in interaction frequency correlates with changes in gene expression.
- To understand how these differences in compartment interaction frequencies relate to interactions with linearly local or distal chromosome regions, the authors calculated the distal-to-local ratio (DLR) of Hi-C interaction frequencies. They concluded that major changes upon constricted migration occur in larger scale chromosome structures.
Global genomic rearrangements occurring after constriction migration
- A translocation between Chr5 and Chr13 was evident only in cells that had undergone constricted migration. The translocation breakpoint falls in a gene poor region of both chromosomes.
- The authors investigated possible copy number variations (CNV) due to constricted migration in all A375 subpopulation of cells. Cells that underwent constricted migration exhibited notable copy number loss in Chr13.
- The authors also observed a decrease in inter-chromosomal fraction of interactions (ICF) of Chr13 in cells that underwent constricted migration.
Metastatic phenotypes in 3D models: collagen matrices
- To investigate whether the cells that had undergone multiple rounds of constricted migration show metastatic behaviour in more biologically relevant environments, the authors embedded embedded cells that either had or had not undergone constricted migration through the Transwell filters in 3D collagen gel matrices.
- Cells that underwent constricted migration displayed long cytoplasmic protrusions and large nuclear deformations, suggesting attempts to migrate through the collagen pores.
- In contrast, cells that did not undergo constricted migration did not exhibit cellular or nuclear shape changes.
Metastatic phenotypes in 3D models: organoid-like conditions
- To investigate the cells’ behaviour in organoid-like conditions, the authors co-cultured each A375 subpopulations of cells with normal human foreskin fibroblasts (BJ-1) on a low adherent surface to induce aggregation and attachment of cells to each other.
- H&E staining of the organoids revealed disorganized aggregations of epithelium clusters in the outside of the organoid, likely arising from the decreased cell-cell attachments and highly migratory phenotypes of the migratory cells.
- Additionally, in the organoids formed by the highly migratory melanoma cells, channel-like openings were observed in the stromal compartment, reminiscent of vasculogenic mimicry.
What I like about this preprint
I found the question the group asked very interesting. I think this work is relevant not only to cancer biology, but to many fields. As a scientist I like taking an inter-disciplinary approach to answer scientific questions, and I think the authors have not only addressed their original question in a multi-disciplinary manner, but they have opened the window into an exciting research area which can potentially benefit from multiple disciplines.
Open questions
- You mentioned in your work some A375 cells that cannot migrate at all. Is this level of heterogeneity similar in other types of cancer cells? Why do you think this population exists? Do they locate at specific sites of your cell culture, for instance suggesting that in an in vivo situation, location within the tumour governs metastatic capacity?
- You mention in your discussion that cells with high capacity for constricted migration have downregulation of genes important for cell-cell and matrix adhesion. Were these genes downregulated since prior to the passage, making these cells already prone to migration?
- Thinking of your and others’ investigations on lamin and heterochromatin, as well as nuclear stability, are there pharmacological agents that can be used with the specific purpose of turning cancerous cells incapable of migrating via effects on lamin and heterochromatin?
- In your work, you mention specific translocation events in chromosomes 5 and 13. Why are these chromosomes affected? Is this the case in other cell types? And why is this of biological relevance in specific contexts (beyond cancer)?
- Your work is fully in the context of cancer pathology, yet many cells are capable of migration. Can you develop further on how could your findings translate to other cell types and non-pathological processes?
References
- Golloshi R, San Martin R, Das P, Raines TI, Thurston D, Freeman T, McCord RP, Constricted migration contributes to persistent 3D genome structure changes associated with an invasive phenotype in melanoma cells, bioRxiv, 2020, doi:10.1101/856583.
Acknowledgement
I thank Rachel Patton McCord and Rosela Golloshi for their engagement and helpful input.
doi: https://doi.org/10.1242/prelights.18058
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cancer biology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Taxane-Induced Conformational Changes in the Microtubule Lattice Activate GEF-H1-Dependent RhoA Signaling
Vibha SINGH
ROCK2 inhibition has a dual role in reducing ECM remodelling and cell growth, while impairing migration and invasion
Sharvari Pitke
Also in the cell biology category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
preLists in the cancer biology category:
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)