A transposase-derived gene required for human brain development
Posted on: 6 July 2023
Preprint posted on 23 May 2023
Article now published in Science Advances at http://dx.doi.org/10.1126/sciadv.adv7530
A domesticated transposase required for healthy neurodevelopment: possibly through developmental apoptosis and synaptic plasticity.
Selected by Amanda IvanoffBackground:
Transposase Co-option
Transposable elements (TEs) cause disease by disrupting host genome structure
(e.g., interrupting or removing host exons, recombining with each other to cause large deletion/duplication events, and misregulating host gene expression) (2). Host defenses chase and repress TEs in an arms race (3, 4). One interesting end for this arms race is co-option: where the host can use this reorganization and introduction of new regulatory (5) or catalytic (6) elements to diversify function of genes and tissues. PiggyBac Transposable Element Derived 5 (PGBD5) is a DNA transposase expressed in most childhood solid tumors where it mediates oncogenic DNA rearrangements (7). Despite this, it is the most conserved domesticated DNA transposase in vertebrates even though it is predominantly expressed in the nervous system, specifically the brain (8). We often assume conservation implies function. Does PGBD5 have a beneficial function in the developing vertebrate brain?
Developmental Apoptosis + DNA Rearrangements
The development of the immune and nervous systems seems uniquely dependent on DNA damage and two of its outcomes: cell apoptosis and DNA rearrangements (1). The potential function of large-scale apoptosis in the developing nervous system is less clear, but one popular hypothesis is that apoptosis maintains synaptic plasticity – and so it is important for memory, learning, and response to injury (9). While genetic mosaicism has been documented, evidence for DNA rearrangements in vertebrate brain development had not been found prior to this preprint (1). Over 50 years ago, it was proposed that protocadherin gene diversification could organize and structure the vertebrate brain (10). In recent years, DNA breaks have been found in many neuronal genes (9). This preprint shows evidence for locus-specific rearrangements in multiple mouse brain tissues which are dependent on the domesticated transposase Pgbd5. Future work might confirm whether factors like Pgbd5 do diversify cell adhesion receptors or other regulators of synaptic connections which organize the vertebrate brain.
Zapater and team analyze the function of PGBD5 in neurodevelopment through:
1) characterizing the effect of Pgbd5’s nuclease activity on brain morphology and behavior in mouse and human
2) confirming whether PGBD5-dependent DNA breaks are repaired and make rearrangements which are sustained to adulthood
Key Findings:
1. Reducing the nuclease activity of Pgbd5 impairs neurodevelopment
Zapater and team use three types of mutations in Pgbd5 to study the effect of the nuclease activity of Pgbd5 on neurodevelopment. First, they found two families through GeneMatcher which carry mutations in PGBD5 near a conserved aspartate triad thought to be necessary for the transposase domain’s biochemical activity. These families had epilepsy, non-progressive motor dysfunction, and impaired expressive language acquisition. Their symptoms paired with thinned corpus collosum and decreased cerebellar volume. Next, they compared phenotypes observed in patients to their own Pgbd5-/- mice (lacking exon 4 of PGBD5). Pgbd5-/- mice similar to the patients had tonic-clonic seizures and reduced cortical and cerebellar volumes. In behavioral tests, Pgbd5-/- mice showed increased locomotor activity, reduced anxiety-like behavior, and impaired motor learning. Comparing mouse behavior to patient symptoms, it seems that PGBD5 has some effect on learning and memory which may be linked to plasticity in development. Last, to confirm if these defects were due to the nuclease activity of Pgbd5, the authors use a knockin mouse model Pgbd5D262A which removes one of the conserved aspartate residues and should reduce Pgbd5 nuclease activity. Strikingly, knockin mice also had tonic-clonic seizures. Based on these three mutations, Pgbd5’s nuclease activity seems necessary for healthy neurodevelopment.
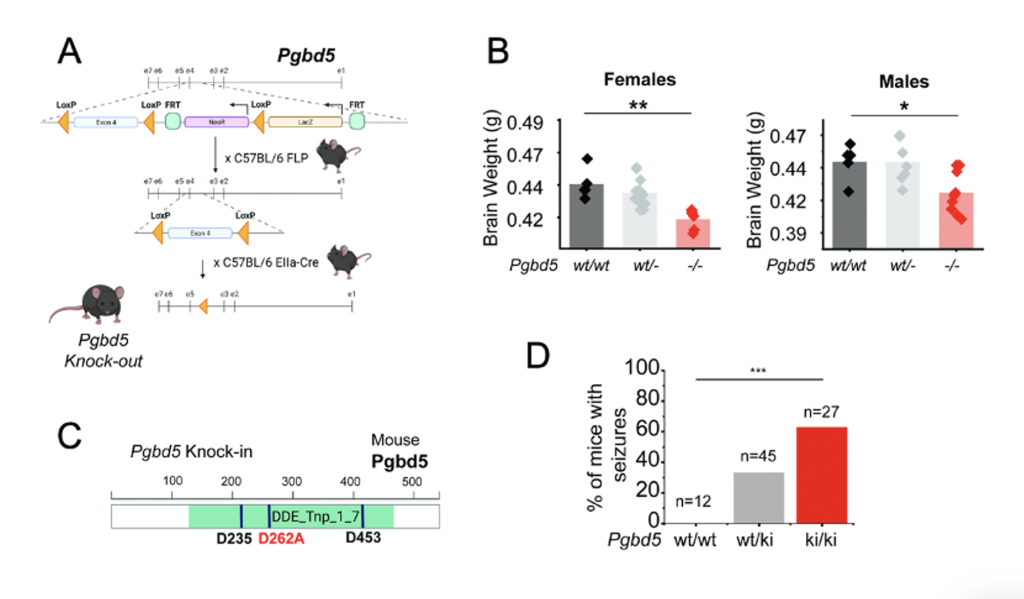
Fig 1. Mutations to PGBD5’s transposase domain cause abnormal brain development in mice
A. Preprint Figure 1H: Schematics of the engineered Pgbd5 locus and breeding strategy to generate Pgbd5-/- mice. B. Preprint Figure 1J: Brain weights of 30-day old WT, Heterozygous, and Pgbd5-/- mice, showing significant reduction in brain weights in male and female mice (t-test p = 0.016 and ** p = 3.4E-3, respectively). Pgbd5wt/wt n=5 female and males, Pgbd5wt/- n=12 females and n=7 males, and Pgbd5-/- n=8 females and n=9 males. C. Preprint Figure 2J: Pgbd5 protein sequence schematic indicating the location of the conserved aspartate triad (black) and the exon 2 D262A substitution (red) D. Preprint Figure 2O: Bar plot of seizure activity Pgbd5-/- versus Pgbd5 D262A/D262A littermate mice (χ2-test p = 7.41E-05). n indicates number of mice assayed.
2. PGBD5 causes DNA rearrangements during cortical development
As neurons exit the cell cycle and specify their cell fate, they accumulate dsDNA breaks and activate end-joining repair. To determine how Pgbd5’s activity is affecting neurodevelopment, the authors focused on the window of peak cortical development in mice (E14.5) and measured the frequency of DNA damage and repair in post-mitotic neurons from WT and Pgbd5 -/- mice. PGBD5 -/- mice had significantly fewer post-mitotic neurons with yH2AX foci at E14.5 than WT mice. This reduction was not observed in mitotic cells nor at E12.5, so PGBD5-dependent breaks seem specific to this stage of development. Next, the authors ask whether PGBD5-dependent breaks require repair and could make sustained rearrangements. They compare break repair in Pgbd5 -/- , Xrcc5 -/- , and double knockout Xrcc5-/- Pgbd5 -/- mice to test whether Xrcc5’s repair mechanism (also used in RAG-mediated DNA rearrangements) can work on Pgbd5-mediated breaks. Pgbd5 -/- mice had fewer post-mitotic neurons with unrepaired breaks at E14.5 than WT mice. Xrcc5-/- mice had more unrepaired breaks (more yH2AX foci) causing increased apoptosis during neurogenesis, but this was not specific to post-mitotic neurons. Xrcc5-/- Pgbd5 -/- mice showed reduced DNA damage only in post-mitotic neurons. Together, these results point to PGBD5-dependent breaks in post-mitotic neurons being repaired by Xrcc5. But are these rearrangements or is this just repair? The authors sampled multiple brain regions from WT and knockout mice for whole genome sequencing to find DNA rearrangements by identifying recurring double-stranded breakpoints or complete overlap regions. WT adult mice had significantly more recurrent DNA rearrangements in multiple brain regions compared to PGBD5 -/- mice. These breakpoints also had a distinct genomic distribution. This trend, combined with previously published evidence of Pgbd5 nuclease activity, indicated Pgbd5 nuclease activity is likely locus-specific.
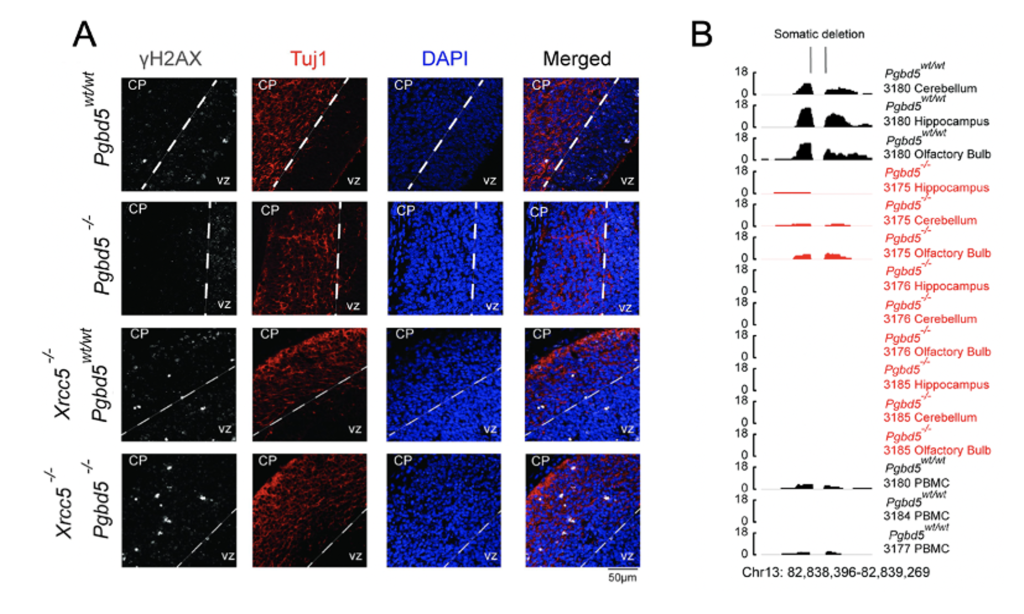
Fig 2. PGBD5-dependent DNA breaks undergo NHEJ repair and form rearrangements sustained into adulthood.
A. Formed from preprint Figures 3A and 4A: Immunofluorescence micrographs of Pgbd5wt/wt , Pgbd5-/-, Xrcc5-/-;Pgbd5wt/wt , and double knockout Xrcc5-/-;Pgbd5-/- 14.5-day old litter mate embryos. DAPI (blue) stains nuclei. γH2AX (white) indicates double-stranded DNA breaks, and Tuj1 (red) stains differentiated post-mitotic neurons. B. Preprint Figure 5E: One histogram of variant junction split and discordant reads used by Delly2 to determine structural variants. Here shown is validated locus chr13:82838396-82839269.
Quick Sum Up and Implications:
Mutations in PGBD5 thought to reduce its nuclease activity cause neurodevelopmental defects in mouse and human. PGBD5-dependent breaks create sustained rearrangements in multiple brain tissues of adult mice. The authors suggest that:
1) PGBD5-dependent DNA breaks may be an underlying mechanism behind developmental apoptosis.
2) PGBD5-dependent DNA rearrangements of target genes may promote neuronal diversification and shape organization of the brain as hypothesized decades ago.
Through both of these mechanisms, and seen in the phenotypes of impaired motor learning and dramatic changes to synaptic strength (namely epilepsy), PGBD5-mediated breaks and rearrangements may also be linked to synaptic plasticity.
Why I chose this preprint:
Nearly half of all cells produced in mammalian neurogenesis die via developmental apoptosis. The evolution of a developmental process that requires such large-scale programmed cell death is incredible in the light of units of selection. Selection would have to act on genes and cells (likely negatively) first before they join to become tissues and organs like the brain, and before any anticipated function for organization in the brain can benefit the organism. I love stories where systems are built despite, or even because of conflicts at lower levels of selection. Similarly, TEs and their host ride the line between friend and foe, and I love studying how cooperation and complexity evolve in systems like this. I think Pgbd5 and factors like it can tell a similar story, by making a mechanistic connection between developmental apoptosis, DNA damage and rearrangements, and synaptic plasticity.
Future Directions/Questions for the Authors:
Q1. How do you think Pgbd5-dependent breaks are biased to post-mitotic neurons?
Or does this more reflect a difference in repair in post-mitotic neurons?
Q2. You mention that the choice to use bulk WGS to identify rearrangements was partly because of the limits on detection of larger variants produced by rearrangements and the requirements of DNA amplification. Is it worth analyzing trends in these breakpoints from the bulk data or looking into expression of alternative transcripts between WT and KO? Have you already seen any functional trends between break sites targeted by Pgbd5 (from either the WGS , ChIP data, or DEG from the knockin model)?
Q3. On this note: Simi et al. posted a preprint (11) shortly after yours where they find higher expression of Pgbd5 at E12 and E18, and show that Pgbd5 seems to gate differentiation and migration of neocortical neurons. Combined with your E14.5 phenotypes, I wondered if either:
-
- This reflects waves where Pgbd5-dependent breaks enable migration through the diversification of target genes.
- This reflects waves where apoptosis gates differentiation of these neurons.
What do you make of these findings?
Cited:
- L. J. Zapater, E. Rodriguez-Fos, M. Planas-Felix, S. Lewis, D. Cameron, P. Demarest, A. Nabila, J. Zhao, P. Bergin, C. Reed, M. Yamada, A. Pagnozzi, C. Nava, E. Bourel-Ponchel, D. E. Neilson, A. Dursun, R. K. Özgül, H. T. Akar, N. D. Socci, M. Hayes, R. Rabadan, D. Torrents, M. C. Kruer, M. Toth, A. Kentsis, A transposase-derived gene required for human brain development (2023), p. 2023.04.28.538770, , doi:10.1101/2023.04.28.538770.
- L. M. Payer, K. H. Burns, Transposable elements in human genetic disease. Nat Rev Genet. 20, 760–772 (2019).
- A. A. Aravin, G. J. Hannon, J. Brennecke, The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–64 (2007).
- F. M. Jacobs, D. Greenberg, N. Nguyen, M. Haeussler, A. D. Ewing et al., An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516:242–45 (2014).
- V. Sundaram, Y. Cheng, Z. Ma, D. Li, X. Xing, P. Edge, M. P. Snyder, T. Wang, Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976 (2014).
- R. L. Cosby, J. Judd, R. Zhang, A. Zhong, N. Garry, E. J. Pritham, C. Feschotte, Recurrent evolution of vertebrate transcription factors by transposase capture. Science. 371, 797 (2021).
- A. G. Henssen et al., PGBD5 promotes site-specific oncogenic mutations in human 45 tumors. Nat Genet 49, 1005-1014 (2017).
- A. G. Henssen, E. Henaff, E. Jiang, A. R. Eisenberg, J. R. Carson, C. M. Villasante, M. Ray, E. Still, M. Burns, J. Gandara, C. Feschotte, C. E. Mason, A. Kentsis, Genomic DNA transposition induced by human PGBD5. eLife. 4, e10565.
- C. P. Gilman, M. P. Mattson, Do apoptotic mechanisms regulate synaptic plasticity and growth-cone motility? Neuromol Med. 2, 197–214 (2002).
- W. R. Gray, W. J. Dreyer, L. Hood, Mechanism of antibody synthesis: size differences between mouse kappa chains. Science 155, 465-467 (1967).
- A. Simi, F. Ansaloni, D. Damiani, A. Codino, D. Mangoni, P. Lau, D. Vozzi, L. Pandolfini, R. Sanges, S. Gustincich, The Pgbd5 DNA transposase is required for mouse cerebral cortex development through DNA double-strand breaks formation (2023), p. 2023.05.09.539730, , doi:10.1101/2023.05.09.539730.
doi: https://doi.org/10.1242/prelights.35009
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the animal behavior and cognition category:
Cannibalism as a mechanism to offset reproductive costs in three-spined sticklebacks
Tina Nguyen
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
Trade-offs between surviving and thriving: A careful balance of physiological limitations and reproductive effort under thermal stress
Tshepiso Majelantle
Also in the developmental biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Also in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
Also in the genetics category:
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Also in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the animal behavior and cognition category:
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Also in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)