Conservation of the Toxoplasma conoid proteome in Plasmodium reveals a cryptic conoid feature that differentiates between blood- and vector-stage zoites
Posted on: 23 July 2020
Preprint posted on 13 July 2020
Article now published in PLOS Biology at http://dx.doi.org/10.1371/journal.pbio.3001081
Categories: cell biology
Background
Apicomplexan parasites are able to actively seek, bind to, and invade the cellular milieu of suitable animal hosts. From there, they manipulate and exploit these host cells to promote their own growth, and their onward transmission. In apicomplexan parasites, the apical complex is key for invasion. The inner membrane complex (IMC) provides shape and protection to the cell, as well as other functions such as gliding motility in apicomplexans. The apical complex has evolved together with the IMC, to provide a location for cellular functions including exocytosis and endocytosis.
The apical complex has been largely studied in ultrastructural studies, and apical rings are the basis of this structure. An apical polar ring (APR) acts as a microtubule organising centre (MTOC) for the subpellicular microtubules, and a second APR coordinates the apical margin of the IMC. Within this opening created by the APRs are further rings, a notable one being the ‘conoid’. The conoidinteracts intimately with secretory organelles including micronemes, rhoptries and other vesicles. While the APRs appear to play key structural organising roles, the conoid is closely associated with the events and routes of vesicular trafficking, delivery and in some cases uptake. While the conoid and its tubulin-based composition have been well described in Toxoplasma, Aconoidasida are considered to have either completely lost the conoid (e.g. Babesia, Theileria), or lost it from multiple zoite stages, e.g. Plasmodium spp. stages other than the ookinete (although controversy exists on whether the conoid was fully lost in Plasmodium spp.). The difficulties that have affected solving this controversy include that relatively few conoid protein and molecular signatures have been identified, making it difficult to conclude whether it is genuinely missing from some major groups, and making it difficult to objectively test for the presence of a homologous structure.
In their work, Koreny et al have carefully explored the Toxoplasma gondii conoid using multiple proteomic approaches, and then explored if these conoid-specific proteins are present in similar locations within the zoite forms (ookinetes, sporozoites and merozoites) of Plasmodium berghei (Fig.1).
Key findings and developments
The authors began by using multiple spatial proteomic methods to identify new candidate conoid proteins. This included the use of the recently published method by the same group, called hyperplexed Localization of Organelle Proteins by Isotope Tagging (hyperLOPIT) (2) over three separate datasets. This allowed assigning 76 proteins to one of the two apical protein clusters, apical 1 and apical 2. These two clusters were verified as comprising proteins specific to structures associated with the conoid, the apical polar ring, and apical cap of the IMC. A further 1013 proteins were quantified in either one or two datasets (instead of the three discussed above), and allowed distinguishing a further 16 proteins assigned to the apical clusters. From all analyses, 92 proteins were assigned as putative apical proteins across the hyperLOPIT samples. While 57 of these proteins had been validated as being located to specific structures (other than the conoid), a remaining 35 protein candidates had had no independent validation on their location.
A second proteomic strategy was used for these candidates, namely, BioID (proximity-dependent biotinylating and pulldown). Two baits used were known conoid markers in Toxoplasma, SAS6-like protein and RNG2. A third bait protein used as negative control, was MORN3, as its location is in high abundance in the apical cap, although excluded in the apex, where the conoid is located. Biotinylated proteins were purified in a streptavidin matrix and analysed by mass spectrometry. Altogether, of the hyperLOPIT-assigned apical proteins, 25 were also detected by BioID with SAS6L and RNG2 but not MORN3. These data indicate that the BioID spatial proteomics indeed enrich for apical proteins, with the differences between SAS6L/RNG2 and MORN3 labelling offering a level of discrimination for conoid-associated proteins when compared to apical cap proteins.
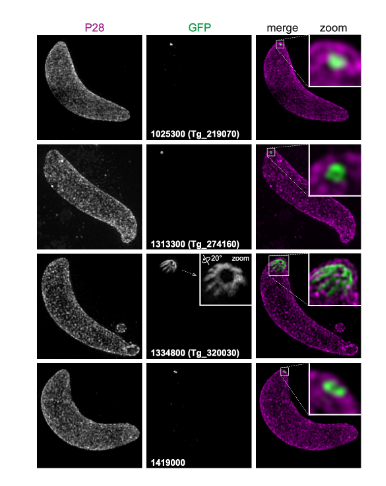
Further validation was performed using fluorescence microscopy. Widefield microscopy showed 13 proteins to be confined in a single small punctum in the extreme apex of the cell. However, higher resolution imaging was required to show specific localization. For this, the authors used 3D-SIM super-resolution to first determine the localizations of SAS6L and RNG2, as these two markers provide definition of the relative positions of the new proteins. Three of the newly identified proteins co-localized to the SAS6L pattern of the full body of the conoid. A further three proteins, with this same pattern of BioID detection, all formed a ring at the posterior end, or base, of the conoid. A further protein was found to co-localize with the RNG2 marker. Altogether, these 3D-SIM data confirm the identities and specific localization of various proteins across various parts of the conoid.
Having validated conoid proteins by various methods in T. gondii, the next step the authors did was to determine how conserved was the conoid proteome in other apicomplexans and related groups within the Myzozoa, and whether there was indeed evidence for conoid loss in the Aconoidasida. They first used two separate bioinformatic/genomic data analysis methods for identification of orthologues. The orthology analysis showed a high degree of conservation (88%) in other coccidia. In other apicomplexan groups (including known members of the Aconoidasida) and in the nearest apicomplexan relatives, the chromerids, approximately half of the conoid proteins are found. The conserved proteins include those associated with all structural components of the T. gondii conoid- conoid body, conoid base, and preconoidal rings.
To test orthologue localization of known T. gondii conoid-associated proteins in Plasmodium, nine selected conoid proteins (representing the conoid’s body and base, and the preconoidal rings) were tagged in P. berghei. This parasite strain provided access to all invasive zoite forms of the parasite: ookinetes, sporozoites, and merozoites. In ookinetes, an apical location was seen for all 9 proteins. In all cases examined, the location and structures formed by the Plasmodium orthologues phenocopied those of T. gondii, strongly suggestive of conservation of function.
With the new markers for components of an apparent conoid in P. berghei, the authors tested for presence of these proteins in the other zoite stages: sporozoites and merozoites. In sporozoites all proteins tested for, were detected in the cell apex. In merozoites, only 6 of the 9 proteins tested were detected, and each formed an apical punctum juxtaposed to the nucleus, consistent with apical location.
What I like about this preprint
Already before I had liked a lot the development of hyperLOPIT that the authors presented. I was wondering on applications, and I was very excited to see it used in this work now. Moreover, I like that they approach questions previously not asked-and challenge assumptions previously made, with new tools, reaching new conclusions. I think this is the exciting part of science! I now look forward to knowing more about the conoid function in the different organisms studied.
References
- Koreny, L., et al Conservation of the Toxoplasma conoid proteome in Plasmodium reveals a cryptic conoid feature that differentiates between blood and vector-stage zoites, bioRxiv, 2020
- Barylyuk, K., A subcellular atlas of Toxoplasma reveals the functional context of the proteome, bioRxiv, 2020.
doi: https://doi.org/10.1242/prelights.23365
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
KANK2 at focal adhesion regulates their maintenance and dynamics, while at fibrillar adhesions it influences cell migration via microtubule-dependent mechanism
Vibha SINGH
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Taxane-Induced Conformational Changes in the Microtubule Lattice Activate GEF-H1-Dependent RhoA Signaling
Vibha SINGH
preLists in the cell biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)