Proteolysis-free cell migration through crowded environments via mechanical worrying
Posted on: 5 December 2022 , updated on: 7 December 2022
Preprint posted on 8 November 2022
“Worry” your way out: Driscoll and colleagues characterize a bleb-based mode of cell migration featuring repetitive agitation of the extracellular matrix.
Selected by Jade ChanCategories: biophysics, cancer biology, cell biology
Background
Migrating cells underlie multiple key processes in health and disease. Two primary modes of migration have been described: mesenchymal and amoeboid. Mesenchymal migration, a “path-making” mode, is characterized by extracellular matrix (ECM) remodelling via matrix metalloproteinase (MMP) secretion and strong cell-substrate interactions through the formation of focal adhesions. Cells undergoing mesenchymal migration often exhibit an elongated shape with well-organized networks of filamentous actin1,2.
In contrast, amoeboid migration, a “path-finding” mode, relies on actomyosin contractility and bleb formation with weak cell-substrate adhesions and minimal degradation of the ECM. Instead of carving out paths via MMP secretion, amoeboid cells deform their bodies to fit through pores in the ECM1-5. Recent studies on melanoma have revealed an enrichment of rounded amoeboid cells at tumour margins with enhanced metastatic capabilities6, 7. Given that most cancer deaths worldwide are associated with metastasis, there is an urgent need to investigate this population of migratory tumour cells.
In this study, Driscoll and colleagues uncover a novel aspect of amoeboid migration wherein cells use polarized blebs to physically degrade ECM fibres in their migratory path. This process of sustained agitation of ECM components is dubbed “worrying.” The authors present a mechanosensitive signalling axis that sustains worrying via adhesion signalling, PI3K/Rac1-mediated actin assembly within large blebs, and matrix degradation that does not rely on MMPs. Importantly, this preprint challenges the pre-existing notion that amoeboid cells do not actively remodel their microenvironment during migration and uses rigorous experimental paradigms to characterize this new type of cell behaviour.
Key Findings
Amoeboid cells create paths through dense ECM without proteolysis
The authors used metastatic melanoma cells to explore bleb-based motility in soft microenvironments. To mimic dense yet soft in vivo conditions, the authors cultured melanoma cells in fibrous collagen I gels at 1 kPa stiffness. The authors then performed 3D light-sheet microscopy on cells migrating through fluorescently labelled collagen networks and measured the size of pores within the network. They observed that while cells had protrusive blebs that were small enough to fit through most pores, their cell bodies and nuclei were often too large, arguing against a deformation-based mode of migration. Despite their large nuclei and cell bodies, these amoeboid melanoma cells were still able to migrate through collagen networks.
Intriguingly, long-term live-cell imaging revealed the presence of tunnels through the collagen matrix, which contrasts previous claims that amoeboid cells do not degrade the ECM. To determine how amoeboid cells were tunnelling, the authors treated the cells with a general MMP inhibitor. Surprisingly, there was no effect on the melanoma cells’ ability to tunnel. Similar results were obtained when the authors treated the cells with a protease inhibitor cocktail. Together, these results indicate that amoeboid melanoma cells remodel their microenvironment to make migratory paths in a proteolysis-independent manner.
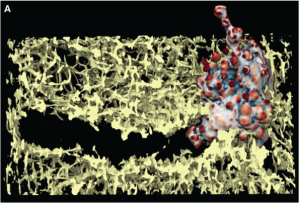
Above: a 3D rendering of a melanoma cell with polarized blebs (coloured by surface curvature) tunnelling through a dense collagen matrix (yellow).
Amoeboid cells make migration paths through dense ECM using large blebs
The authors wondered how amoeboid cells were tunnelling without using proteases. Analysis of live-cell imaging videos revealed that cells within tunnels were highly polarized with respect to bleb position and size, with larger blebs located at the cell front. The authors also noted that collagen fibres were often fragmented in proximity to blebs. Using a 3D optical flow algorithm to analyze the motion of collagen strands near protrusive and retractive blebs on migrating cells, they observed that collagen near the front of highly polarized cells underwent prolonged agitation and internalization via macropinocytosis. Overall, these observations suggest that amoeboid cells use large, polarized blebs to physically ablate collagen fibres in their path.
PI3K activity governs bleb size in migrating cells
The authors sought to uncover molecular players involved in maintaining bleb size and polarity. Staining of the focal adhesion marker paxillin revealed that blebs contacted collagen fibres through adhesion complexes. Strikingly, PI3K, a cell polarity factor recruited by focal adhesion kinase (FAK)8-10, was also enriched in large blebs at the cell front. Adhesions formed by large blebs were longer-lived than nascent adhesions of cells cultured on coverslips11, suggesting that the location and persistence of adhesions near the migrating cell front recruits PI3K. To confirm this, the authors treated the cells with a small molecule FAK inhibitor. While the bleb number and polarization were unaffected by FAK inhibition, the overall volume of blebs was decreased. Furthermore, when examining the molecular events occurring after FAK inhibition, the authors found that PI3K polarization was abolished followed by reduced bleb volume. To test this causal link directly, they used an optogenetic strategy to enhance PI3K activity with high spatial specificity in migrating cells. Indeed, PI3K photoactivation induced local bleb swelling, whereas treatment with a pharmacological PI3K inhibitor resulted in bleb shrinkage.
PI3K increases bleb size via branched actin polymerization
How does PI3K activity promote bleb growth? Prior studies have suggested that PI3K activity is coupled with actin protrusion, a phenomenon typically associated with mesenchymal rather than amoeboid migration. In contrast, bleb expansion in amoeboid cells is attributed to membrane extrusion driven by cytoplasmic pressure12. To explore this apparent contradiction, the authors performed total internal reflection fluorescence (TIRF) imaging in live cells expressing beta-actin-GFP and observed that actin filaments assembled inside blebs during expansion. These observations support the hypothesis that bleb growth can also be driven by actin polymerization.
To link PI3K activity with actin filament assembly and bleb growth, the authors investigated the Rac1 – WAVE – Arp2/3 signalling axis, which is active during lamellipodia expansion13,14. Treating migrating cells with CK666, an Arp2/3 inhibitor, decreased bleb volume. Furthermore, using a light-sensitive Rac1 expression construct, the authors demonstrated that Rac1 photoactivation increased bleb size in a reversible manner. For greater spatial control, they also performed similar experiments using a light-sensitive Tiam1 (a guanine nucleotide exchange factor that specifically acts on Rac1) construct that could be uncaged and sequestered in specific intracellular locations. Consistent with previous results, local Tiam1 activation significantly increased bleb volume, as well as Arp2/3 concentration within the expanding blebs. Altogether, these experiments uncover the connection between PI3K activity and actin-based bleb expansion in “worrying” cells.
Why I chose this preprint
I was highly interested in this preprint because of the characterization of a novel type of cell behaviour during migration. The finding that amoeboid cells physically manipulate and ablate the ECM via mechanical “worrying” challenges the canonical view of amoeboid migration. Further detailed analyses of these behaviours could potentially lead to the development of therapeutic strategies specifically targeting metastatic tumour cells that do not rely on MMPs or other proteases to invade healthy tissue. It would be fascinating to determine whether non-tumoural cell types that undergo amoeboid migration (such as leukocytes) “worry” in normal physiological contexts, or if this behaviour is specific to tumour cells.
In my own research, I am currently investigating a protein that appears to regulate mesenchymal versus amoeboid migration in glioblastoma (GBM), a highly invasive brain tumour. While GBM cells are thought to undergo mesenchymal migration, tissue stiffness within tumours is highly heterogeneous, which may promote different types of migration. It would be interesting to investigate whether invasive GBM cells also exhibit “worrying” behaviour at tumour margins.
Questions for the authors
- Aside from PI3K’s well-established role in growth factor signalling that contributes to tumorigenesis, is there any clinical or experimental evidence suggesting that tumours with PI3K overexpression or hyperactivity are more metastatic than tumours with wild type PI3K?
- Is PI3K activity or expression spatially heterogeneous in solid tumours?
- In your live-cell imaging experiments, did you observe any cells that exhibited plasticity between the different migration modes (i.e. cells that switched between deformation-based amoeboid migration, “worrying” amoeboid migration, and/or mesenchymal migration)?
References
- Yamada, K. M., & Sixt, M. (2019). Mechanisms of 3D cell migration. Nature reviews. Molecular cell biology, 20(12), 738–752. https://doi.org/10.1038/s41580-019-0172-9
- Sahai, E., & Marshall, C. J. (2003). Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nature cell biology, 5(8), 711–719. https://doi.org/10.1038/ncb1019
- Ruprecht, V., Wieser, S., Callan-Jones, A., Smutny, M., Morita, H., Sako, K., Barone, V., Ritsch-Marte, M., Sixt, M., Voituriez, R., & Heisenberg, C. P. (2015). Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell, 160(4), 673–685. https://doi.org/10.1016/j.cell.2015.01.008
- Lämmermann, T., Bader, B. L., Monkley, S. J., Worbs, T., Wedlich-Söldner, R., Hirsch, K., Keller, M., Förster, R., Critchley, D. R., Fässler, R., & Sixt, M. (2008). Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature, 453(7191), 51–55. https://doi.org/10.1038/nature06887
- Wolf, K., Te Lindert, M., Krause, M., Alexander, S., Te Riet, J., Willis, A. L., Hoffman, R. M., Figdor, C. G., Weiss, S. J., & Friedl, P. (2013). Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. The Journal of cell biology, 201(7), 1069–1084. https://doi.org/10.1083/jcb.201210152
- Orgaz, J. L., Crosas-Molist, E., Sadok, A., Perdrix-Rosell, A., Maiques, O., Rodriguez-Hernandez, I., Monger, J., Mele, S., Georgouli, M., Bridgeman, V., Karagiannis, P., Lee, R., Pandya, P., Boehme, L., Wallberg, F., Tape, C., Karagiannis, S. N., Malanchi, I., & Sanz-Moreno, V. (2020). Myosin II Reactivation and Cytoskeletal Remodeling as a Hallmark and a Vulnerability in Melanoma Therapy Resistance. Cancer cell, 37(1), 85–103.e9. https://doi.org/10.1016/j.ccell.2019.12.003
- Georgouli, M., Herraiz, C., Crosas-Molist, E., Fanshawe, B., Maiques, O., Perdrix, A., Pandya, P., Rodriguez-Hernandez, I., Ilieva, K. M., Cantelli, G., Karagiannis, P., Mele, S., Lam, H., Josephs, D. H., Matias-Guiu, X., Marti, R. M., Nestle, F. O., Orgaz, J. L., Malanchi, I., Fruhwirth, G. O., … Sanz-Moreno, V. (2019). Regional Activation of Myosin II in Cancer Cells Drives Tumor Progression via a Secretory Cross-Talk with the Immune Microenvironment. Cell, 176(4), 757–774.e23. https://doi.org/10.1016/j.cell.2018.12.038
- Chen, H. C., & Guan, J. L. (1994). Association of focal adhesion kinase with its potential substrate phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America, 91(21), 10148–10152. https://doi.org/10.1073/pnas.91.21.10148
- Johnson, H. E., King, S. J., Asokan, S. B., Rotty, J. D., Bear, J. E., & Haugh, J. M. (2015). F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. The Journal of cell biology, 208(4), 443–455. https://doi.org/10.1083/jcb.201406102
- Welf, E. S., Ahmed, S., Johnson, H. E., Melvin, A. T., & Haugh, J. M. (2012). Migrating fibroblasts reorient directionality by a metastable, PI3K-dependent mechanism. The Journal of cell biology, 197(1), 105–114. https://doi.org/10.1083/jcb.201108152
- Choi, C. K., Vicente-Manzanares, M., Zareno, J., Whitmore, L. A., Mogilner, A., & Horwitz, A. R. (2008). Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nature cell biology, 10(9), 1039–1050. https://doi.org/10.1038/ncb1763
- Cunningham C. C. (1995). Actin polymerization and intracellular solvent flow in cell surface blebbing. The Journal of cell biology, 129(6), 1589–1599. https://doi.org/10.1083/jcb.129.6.1589
- Welch, H. C., Coadwell, W. J., Stephens, L. R., & Hawkins, P. T. (2003). Phosphoinositide 3-kinase-dependent activation of Rac. FEBS letters, 546(1), 93–97. https://doi.org/10.1016/s0014-5793(03)00454-x
- Bisi, S., Disanza, A., Malinverno, C., Frittoli, E., Palamidessi, A., & Scita, G. (2013). Membrane and actin dynamics interplay at lamellipodia leading edge. Current opinion in cell biology, 25(5), 565–573. https://doi.org/10.1016/j.ceb.2013.04.001
doi: https://doi.org/10.1242/prelights.33209
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
Also in the cancer biology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Taxane-Induced Conformational Changes in the Microtubule Lattice Activate GEF-H1-Dependent RhoA Signaling
Vibha SINGH
ROCK2 inhibition has a dual role in reducing ECM remodelling and cell growth, while impairing migration and invasion
Sharvari Pitke
Also in the cell biology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cancer biology category:
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Also in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (1 votes)
(1 votes)