A non-canonical arm of UPRER mediates longevity through ER remodeling and lipophagy.
Posted on: 17 December 2018
Preprint posted on 17 December 2018
A neuronal stress response mediates longevity via cross-tissue signalling and upregulation of lipophagy
Selected by Sandra Malmgren HillCategories: cell biology, genetics, molecular biology
Context
Aging is defined as a time-dependent increase in mortality, and is thought to be caused by a loss of homeostasis and systemic collapse due to the build-up of harmful material1. As damaged components such as dysfunctional proteins and organelles accumulate in the cells of an aging organism, this induces a range of genetic responses in an attempt to combat the increase in stress. It has been shown that stress responses between different organelles in the cell are interconnected to provide increased buffering and robustness2. However, little is known about how cells in different tissues of an organism are connected and can signal to each other to warn about impeding danger. In a previous study, the authors of this preprint showed that in C. elegans, the accumulation of misfolded proteins in the endoplasmic reticulum (ER) of neurons induces an unfolded protein response (UPRER) that is transmitted throughout the organism to yield a similar response in cells of the intestine3. The group showed that cells of the intestine responded by inducing the heatshock response, and that this cell non-autonomous signalling counteracted the age-related loss of protein quality control4, and improved longevity.
In the study highlighted by this prelight, the authors expand on their findings and present data that the induction of heatshock response might not be the whole story. They show that the UPRER induced signal from neurons causes a dramatic remodelling of ER and lipid content of intestinal cells and that these events, rather than the induced heatshock response, are responsible for the observed lifespan extension.
Major findings
The authors use a long-lived model of the nematode C. elegans (median age 24 days compared to 20 days in wildtype), which harbours a constitutively active unfolded response of the ER (UPRER) in its neuronal cells. This is achieved by tissue specific overexpression of the spliced version of the transcription factor XBP-1, bypassing the need of IRE-1 activation for UPR induction5. The authors then study the cell non-autonomous response in the cells of the intestine and identify a striking morphological change of the ER in early adulthood of these animals. Using a fluorescent marker of the ER lumen, the authors describe the formation of circular ER-derived membranes (CERMs) in the intestine. These structures are only seen in animals with neuronal xbp-1 overexpression and are visible only at day 4 of adulthood, after which they seem to disappear. Furthermore, the formation of these CERMs is dependent on expression of xbp-1 also in the peripheral tissue, but does not require the induction of the canonical UPR response. Interestingly, CERM formation is not observed when using a systemic overexpression of xbp-1, although these animals still overexpress xbp-1 in neurons, indicating that there might be some kind of feedback signalling.
Using a sorter based LAMPro technology6 the authors were able to analyze whole-body neutral lipid contents and found that the neuronal induction of UPRER caused drastic lipid depletion in intestinal cells. This lipid depletion was explained by an increase in intestinal secretion and degradation of lipid droplets (LDs) by lipophagy. In agreement with an increase in lipophagy upon xbp-1 overexpression, the authors found an increase in the number of lysosomes in the intestinal cells, and they identified the RAB-10/EHBP-1/RME-1 lipophagy complex as being essential for lipid depletion and lifespan extension. This complex was however not essential for the induction of UPRER in the intestine, demonstrating that the effects on ER-remodelling and lipophagy can be uncoupled from the canonical heatshock.
Taken together, the data presented in this pre-print highlight a novel mechanism of cross-tissue signalling, where a neuronal stress response mediates longevity via tissue-specific upregulation of lipophagy.
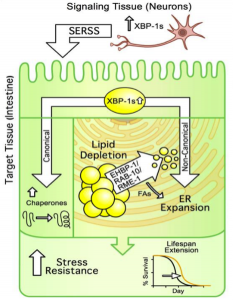
From figure 5 of the pre-print: Upon constitutive induction of UPRER in neurons, mediated by xbp-1 overexpression, a small ER-stress signal (SERSS) is transmitted to the cells of the intestine. This induces a canonical heatshock response, with upregulation of several chaperones, but also leads to a non-canonical response with ER remodelling and EHBP-1 dependent lipophagy. These cell non autonomous responses to UPRER leads to increased stress resistance and longevity.
Why I chose this pre-print
Research on the molecular mechanisms of aging is of great importance, as it is the most predominant risk factor for diseases that compromise human healthspan. While most studies use a reductionist approach, studying one process in one cell/tissue to determine its role in aging, this study uses a more holistic approach, looking at the aging process from the organismal point of view. I find this angle very interesting, and it is intriguing to discover how events in one tissue might influence the response in others, and how this impacts the process of aging. While the mechanisms underlying the observations in this study remain to be identified, I believe that the type of interconnectivity described in this pre-print is of great interest to help us understand how biological systems work, and how they might collapse.
Open questions
- In the previous paper describing signalling between neurons and cells of the intestine the authors showed that the signalling was dependent on Unc-13 mediated release of small clear vesicles. It remains to be determined if the events described in this pre-print are mediated by the same signaling molecule, and a major open question is of course: what is the identity of this cross-tissue signaling molecule? Are xbp-1 overexpressing neurons releasing a different set of neurotransmitters compared to neurons from a wildtype animal?
- The ER-remodelling events described by the authors are present only at day 4 of the adult animal, and then disappears as the animal ages, yet these changes are enough to extend the lifespan of the animal. What are the downstream mechanisms of the described events that ultimately lead to increased stress resistance and longevity?
- The function and nature of the circular ER-derived membranes (CERMs) remain unclear, and it is still to be determined whether these structures are identical to the ER “whorls” observed in ER-phagy or if these structures are a novel type of ER-derived vesicles. Could these structures perhaps be the result of a degradation pathway to remove selective parts of the ER membrane, similar to what has been described for mitochondria?7
- The data in this paper describe a cell non-autonomous response in intestinal cells upon neuronal overexpression of a protein. It will be of interest to determine the physiological relevance for the described neuron-to-intestine signalling. Are similar cross-tissue responses seen during normal aging, or when the animal encounters stress? What other types of cross-tissue interactions are occurring and what are the consequences and the importance of this type of signalling?
References
- Rose MR. Evolutionary biology of aging. Oxford University Press: New York, 1991.
- Dillin A, Gottschling DE, Nystrom T. The good and the bad of being connected: the integrons of aging. Curr Opin Cell Biol 2014, 26: 107-112.
- Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 2013, 153(7): 1435-1447.
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A 2009, 106(35): 14914-14919.
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90(6): 1031-1039.
- Daniele JR, Esping DJ, Garcia G, Parsons LS, Arriaga EA, Dillin A. “High-Throughput Characterization of Region-Specific Mitochondrial Function and Morphology”. Sci Rep 2017, 7(1): 6749.
- Hughes AL, Hughes CE, Henderson KA, Yazvenko N, Gottschling DE. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. Elife 2016, 5.
doi: https://doi.org/10.1242/prelights.6497
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Cell cycle-dependent mRNA localization in P-bodies
Mohammed JALLOH
Control of Inflammatory Response by Tissue Microenvironment
Roberto Amadio
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the genetics category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Lipid-Based Transfection of Zebrafish Embryos: A Robust Protocol for Nucleic Acid Delivery
Roberto Rodríguez-Morales
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
María Mariner-Faulí
Also in the molecular biology category:
Cell cycle-dependent mRNA localization in P-bodies
Mohammed JALLOH
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Fetal brain response to maternal inflammation requires microglia
Manuel Lessi
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the genetics category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)