A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance
Preprint posted on 22 February 2019 https://www.biorxiv.org/content/10.1101/529222v2
Article now published in The Journal of Cell Biology at http://dx.doi.org/10.1083/jcb.201902088
Helping neurons find the other side of the brain: two ubiquitin ligases play an unexpected, non-degradative role at cell tips.
Selected by Angika BasantCategories: cell biology, neuroscience
Background: The leading edge of a migrating cell or the growth cone of an axon comprises a dense meshwork of short actin filaments which form protrusive structures called lamellopodia. A cell actively sensing its environment can also generate longer, exploratory finger-like projections called filopodia. Filopodia emerge from the actin meshwork in the lamellopodia and a key protein required for their formation is VASP (first identified in Drosophila as Enabled (Ena)) (1,2). VASP is an actin assembly factor that competes with capping proteins (that otherwise restrict the length of actin filaments in lamellopodia). Mechanistic details of how VASP does its job is of much interest to cell biologists. For example, we have learnt many details of how it achieves high processivity and cooperates with actin bundling proteins (3, 4) to create the elongated actin structures filopodia are composed of.
Here, I highlight a recent preprint that reveals an interesting role for two ubiquitin (Ub) ligases in VASP regulation, in the context of the developing mouse brain. The Gupton lab has previously demonstrated a role for a Ub ligase TRIM9 in axon branching and filopodia dynamics (5) and has also characterised defects in behaviour and neuronal anatomy of mice lacking another ligase, TRIM67 (6). This preprint illustrates how these two ligases can functionally interact to control VASP activity in growing axons. These studies also expand our view of non-degradative functions of ubiquitinylation in cell biology and signalling (7-9).
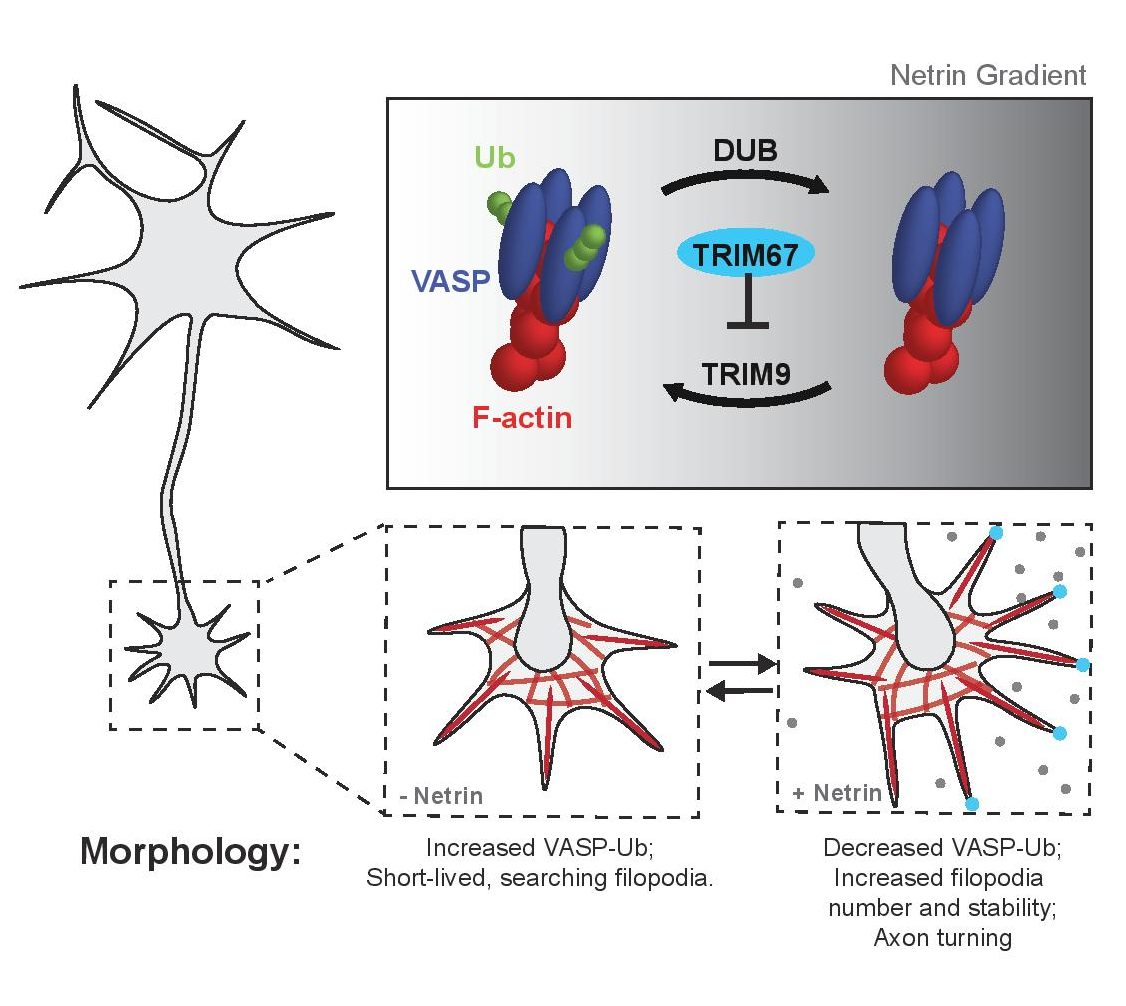
Key findings: Examining sections of the perinatal Trim67-/- mouse brain, the authors find delays in development of the corpus callosum, which is a tract of nerve fibres that connect the two hemispheres of the brain. Specifically, the leading front of these fibres shows larger gaps between the hemispheres in the absence of TRIM67. Cultured neurons from this mutant show obvious defects in their ability to change axon direction in response to the extracellular cue netrin. Trim67-/- neurons in culture also do not show extensive axonal branching in the presence of netrin.
The authors also describe other cell biological and morphological changes in Trim67-/- neurons in detail, such as growth cone morphologies by scanning electron microscopy and measurement of filopodia lifetime by live cell imaging. They find that Trim67-/- neurons spend less time retracting their filopodia in response to netrin. However, these latter changes are complex and may be challenging for a non-expert to appreciate.
TRIM67 is a large multi-domain protein comprising N-terminal tripartite motif domains – a RING domain that confers ligase activity, a B-box domain and a coiled coil domain. Additionally, it contains a COS domain and an FN3 domain that may allow interaction with microtubules and a SPRY domain which typically engages other binding partners. Performing structure-function analysis by expressing versions lacking individual domains, the authors find that all domains participate in the axon-branching response to netrin.
To test whether TRIM67 regulates VASP via a direct interaction, the authors overexpressed constructs of TRIM67 and VASP in 293T cells. They found that these proteins interact. The coiled coil domain of TRIM67 is required for this interaction but its ligase activity is not. Live TIRF microscopy of axonal filopodia suggests that colocalization of TRIM67 and VASP is reduced without the coiled coil domain.
Rather interestingly, both TRIM9 and TRIM67 interact with the VASP EVH1 domain. TRIM9 was previously shown to ubiquitinate VASP (5). In TRIM67-/- neurons, VASP-Ub levels were increased. Epistasis experiments in this study reveal that the Trim9-/-;Trim67-/- double knockout resembles Trim9-/- in that relative levels of VASP-Ub are lower and number of filopodia per growth cone follows the same trend as Trim9-/-. This suggests that TRIM67 acts in the same pathway as TRIM9 and likely inhibits activity of the latter towards VASP via competitive binding. The combined function of these two Ub ligases contributes to filopodia formation and axon guidance.
What I like about this preprint: This paper and others from the Gupton lab combine a variety of approaches – biochemical analyses, live cell imaging and an in vivo animal model – to reveal an unexpected pathway of VASP regulation. This study also opens up many interesting new questions for enthusiasts of cytoskeletal regulation.
Future directions and questions for the authors:
As one of the aforementioned enthusiasts of cytoskeletal regulation (and not an expert in ubiquitinylation!), I’d love to hear the authors’ thoughts on how exactly TRIM9 and TRIM67 would exert their function. Specifically:
- How does the presence of netrin reverse VASP Ub or how are DUBs activated?
- What contributes to increased VASP-Ub in TRIM9-/- neurons in response to netrin (Figure 5F)? Is there a redundant Ub-ligase?
- Why is the ligase activity of TRIM67 required to rescue VASP-Ub in 293T cells (Figure 5G-H)?
- Can TRIM9 and TRIM67 heterodimerize?
- Would you expect TRIM67, TRIM9 and VASP to form a stable or transient ternary complex? Would DCC be part of such a complex?
- How long are the Ub chains that TRIM9 would typically attach to VASP and would this matter to the mechanism?
References:
- Chesarone and Goode, Op. in Cell Biol 2009
- Faix et al., Int J Biochem Cell Biol 2009
- Winkelman et al., PNAS 2014
- Harker et al., MBoC 2019
- Menon et al., Dev Cell 2015
- Boyer et al., eNeuro 2018
- Dwane et al., JBC 2017
- Ball et al., PLoS Comp Biol 2016
- O’Neill JBC 2008
Posted on: 28 February 2019 , updated on: 6 March 2019
doi: https://doi.org/10.1242/prelights.9083
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Also in the neuroscience category:
Fetal brain response to maternal inflammation requires microglia
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the neuroscience category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)