Interspecies differences in proteome turnover kinetics are correlated with lifespans and energetic demands
Posted on: 18 May 2020
Preprint posted on 27 April 2020
Article now published in Molecular & Cellular Proteomics at http://dx.doi.org/10.1074/mcp.RA120.002301
Live long and proteostasis: slower protein turnover correlates with a longer lifespan and reduced energy consumption.
Selected by Teresa RayonCategories: biochemistry, cell biology, evolutionary biology
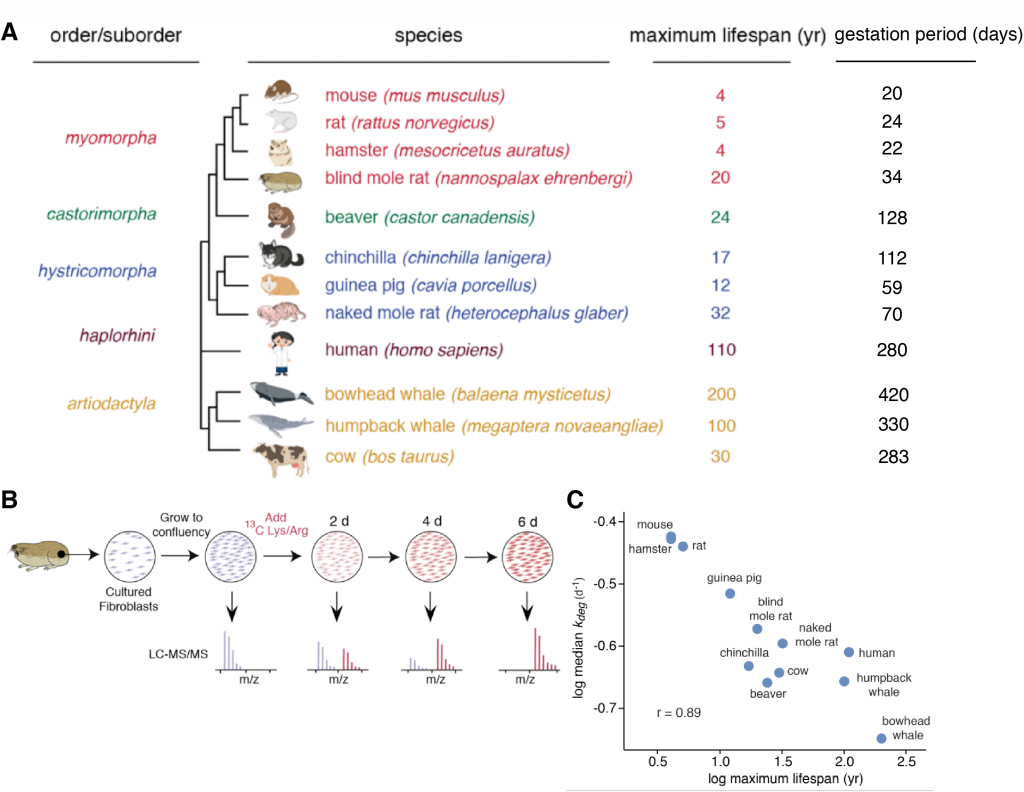
Protein homeostasis (proteostasis) is essential for an organism to maintain its normal cellular function, as cells need to continually degrade and replace damaged and old proteins. Proteostasis ensures timely disposal of misfolded proteins and the correct balance between protein production and degradation (i.e protein turnover) As we age, the capacity of cells to maintain proteostasis declines progressively. In addition, several models for lifespan extension in mammalian systems have shown reduced protein turnover rates (Basisty et al., 2018). But is enhanced protein quality and reduced protein turnover the underlying mechanism behind longevity? In this preprint, Swovick et al. measure protein turnover kinetics across twelve different mammals with lifespans ranging from 4 years in mouse to 200 years in the bowhead whale to answer this question.
In a tour de force, the authors measure degradation rates of single proteins genome-wide by using quantitative proteomics (pulsed Stable Isotope Labeling in Cell Culture -pSILAC-) of quiescent fibroblasts. They find that longer-lived organisms generally have slower global protein turnover rates. This interesting correlation drives the authors to ask why long-lived organisms have slower rates of protein turnover. They perform an in-depth comparison between the mouse and the evolutionarily related and similar sized naked mole-rat, which has cancer resistance and a long lifespan and therefore is broadly used as a model in the aging field. Through a number of experiments to measure energy consumption and response to proteotoxic stress, they find that increased protein stability reduces ATP demands and reduces the production of reactive oxygen species (ROS) over the course of a long lifespan.
Why I chose the paper:
I have been wondering for a while why developmental pace differs across species. The gestational period is highly species-specific, lasting 420 days in bowhead whales, 280 days in humans, 70 days in naked mole rats and 20 days in mouse, and correlates with the animals’ lifespan. In our own recent work, we find that interspecies differences in protein stability correlate with developmental pace (Rayon et al., 2019). Interestingly, the preprint highlighted here studies the relationship between lifespan -total length of time from birth to death- and protein turnover, and it describes a correlation between protein stability and lifespan in the fibroblasts of 12 different mammalian species. Only recently, advances in quantitative proteomics have enabled the genome-wide measurement of protein turnover rates for individual proteins. I find the use of this state-of-the-art technique to compare between quiescent fibroblasts from multiple species really elegant. This is an impressively large cross-species comparison that includes amazing mammalian species such as the bowhead whale and the naked mole-rat. Moreover, their findings go beyond a correlation of protein stability and lifespan to show that highly abundant proteins in long-lived organisms are significantly more stable. Since protein turnover is highly expensive energetically (Ramsey et al., 2000), this underscores the fact that the cost of proteostasis might be a target for longevity. In agreement with this, they also demonstrate slower rates of ATP production and reduced levels of respiration and glycolytic proteins in long-lived naked mole-rat fibroblasts. Finally, their comparison between mouse and naked mole-rat allows them to show a lower accumulation of ROS in the latter, suggesting that longevity might be related to a reduction in energy demand and ROS production.
Aging is an intrinsic decline in physiological function, that is thought to be due to direct damage, accumulation of cellular waste, errors, and imperfect repairs. Thus, it could be argued that aging starts during development, and it might suggest that proteostasis is a common cellular clock that tracks the course of time during development and adulthood.
Why I think this work moves the field forward
The quest for the elixir of youth is linked to the idea that most of us want to live long, or age slowly, if at all. Faster protein turnover is generally associated with youth, and the progressive loss of proteostasis is a hallmark of aging. Constitutive protein turnover accounts for as much as 25% of the total energy expenditure in the body (Ramsey et al., 2000), and the findings by Swovick et al. support that slower protein turnover rates reduce ATP demand and lower the production of ROS over the course of a long lifespan. Altogether, this work establishes how the energetic costs in protein turnover might determine longevity in mammals. In the future, understanding how long-lived animals ease aging by studying the relationship between metabolism and proteostasis may shed some light towards finding the elixir of youth to extend the lifespan in humans.
Questions to authors
- Fibroblasts senesce and lose amplification potential over passages in culture, and it is thought that the age of the donors’ fibroblasts determines the efficiency of reprogramming. Did the authors take into account the age of the donor species for the isolated fibroblasts and the number of passages of the fibroblasts? I wonder if there would be identifiable differences between old-donor versus young-donor fibroblasts across species.
- Unlike most mammals, naked mole rats do not regulate their body temperature. Do the authors think that the energetic cost and the mechanisms to lower ROS production to maintain a slower protein turnover in other long-lived species would be conserved? Do they think temperature can have an effect on proteostasis?
- In this work the authors measure degradation rates of long-lived proteins. Do the authors think that they would find differences if they measured the stability of short-lived proteins?
- The authors test the ability of naked mole rat and mouse fibroblasts to tolerate proteotoxic stress. Have the authors considered looking separately at the unfolded protein response or the heat shock response?
- The authors measured protein turnover in quiescent cells, what do the authors think would be the effect of cell proliferation rate?
References
Basisty, N., Meyer, J. G. and Schilling, B. (2018). Protein Turnover in Aging and Longevity. PROTEOMICS 18, 1700108.
Ramsey, J. J., Harper, M. E. and Weindruch, R. (2000). Restriction of energy intake, energy expenditure, and aging. Free Radical Biology and Medicine 29, 946–968.
Rayon, T., Stamataki, D., Perez-carrasco, R., Garcia-perez, L., Barrington, C., Melchionda, M., Exelby, K., Tybulewicz, V., Fisher, E. M. C. and Briscoe, J. (2019). Species-specific developmental timing is associated with global differences in protein stability in mouse and human. bioRxiv.
doi: https://doi.org/10.1242/prelights.20619
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
14-3-3 binding regulates Tau assembly and microtubule association
Barbora Knotkova et al.
Structural basis of respiratory complexes adaptation to cold temperatures
Pamela Ornelas
Also in the cell biology category:
Cell cycle-dependent mRNA localization in P-bodies
Mohammed JALLOH
Control of Inflammatory Response by Tissue Microenvironment
Roberto Amadio
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the evolutionary biology category:
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Fetal brain response to maternal inflammation requires microglia
Manuel Lessi
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the evolutionary biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)