Minimal membrane interactions conferred by Rheb C-terminal farnesylation are essential for mTORC1 activation
Posted on: 5 February 2019 , updated on: 11 March 2019
Preprint posted on 8 January 2019
Article now published in Molecular Biology of the Cell at http://dx.doi.org/10.1091/mbc.E19-03-0146
Categories: cell biology
Context
The mechanistic target of rapamycin complex 1 (mTORC1) is a serine/threonine kinase complex that functions as a major regulator of cell growth and metabolism in response to energy levels and nutrient abundance1. In the presence of nutrients, mTORC1 is recruited to the lysosomal surface by RAPTOR and activated Rag GTPases. Once on the lysosomal membrane mTORC1 is activated in parallel by the Rag GTPases together with the Rheb GTPase. The Rheb GTPase itself constitutes a point of regulation as its activity is governed by the TSC1/TSC2 complex which drives the hydrolysis of GTP to GDP in Rheb, thereby causing its inactivation2. The activity of TSC1/TSC2 and thereby mTORC activation by Rheb is regulated by a plethora of signals from environmental cues including growth factor presence and energy levels3 (Figure 1).
While much is known about the upstream signaling that converges on mTORC1 localization and activation, it is not fully established how Rheb localization is regulated. It has been suggested that Rheb localization to endosomes and/or lysosomes is mediated by farnesylation of the C-terminal CaaX motif4. Other studies question the specificity and strength of this localization signal, and suggest that Rheb resides at various endomembrane structures, including the ER and Golgi5. In the case of Rheb residing on Golgi membranes, it is proposed that close proximity with mTORC1 on lysosomal membranes is mediated via interconnected surfaces between these organelles6.
In this preprint, the authors attempt to shed further light on the regulation of Rheb localization and provide evidence for a mechanism where farnesylation of Rheb recruits it to ER membranes where it mediates its mTORC1 activation effect.
Major findings
In this preprint, the authors use immunofluorescence and statistical analysis of colocalization to show that Rheb is not localized to lysosomes, in contrast to what has been published previously. They explain that the overlap with lysosomal LAMP1 is no greater than chance, and that the broad distribution of Rheb in the cell inevitably causes some degree of overlap. The authors confirm this data by showing the same localization of endogenously expressed Rheb carrying an N-terminal HA-tag. In addition, starvation followed by refeeding does not induce a relocalization of Rheb, as is seen for mTORC1 which relocalizes to lysosomes
Utilizing exogenous expression of GFP-tagged Rheb, the authors noticed a network-like distribution. They found that this signal overlapped with the ER-marker Sec61, indicating that Rheb might localize to ER tubules. Furthermore, lysosomes were frequently observed close to Rheb positive ER-structures, providing evidence that ER-resident Rheb could be positioned in close proximity to mTORC1 on lysosomes.
Deletion of the CaaX farnesylation motif inhibited ER localization of Rheb, and also abolished mTORC1 activation. The authors then studied the activity of different hybrid versions of Rheb, constitutively targeted to the plasma membrane or to the ER. Interestingly, constitutive targeting of Rheb to the ER reduced its ability to activate mTORC1, even more so than did constitutive targeting of Rheb to the plasma membrane.
Subcellular fractionation experiments revealed that Rheb is detected mainly in in the cytosolic fraction, evident of only weak interactions with membranes. Thus the authors conclude that farnesylation of Rheb enables its weak interaction with the ER membrane and that this transient and reversible interaction is essential for the interaction with, and activation of, mTORC1.
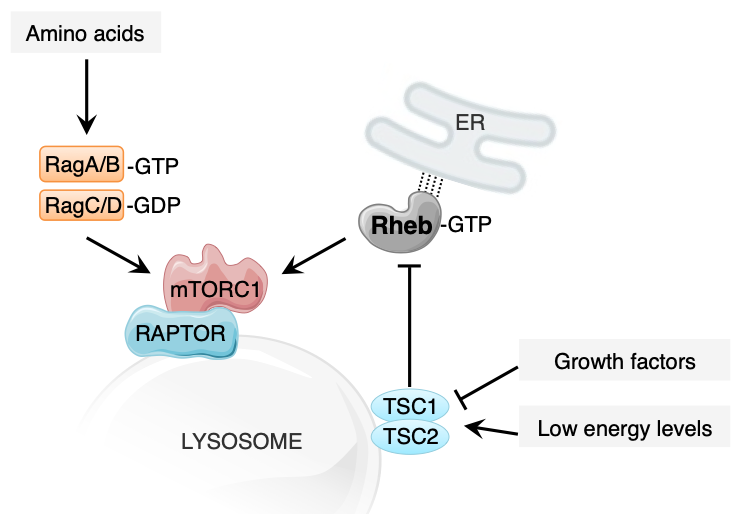
FIGURE 1. Simplified overview of the recruitment and activation of mTORC1 on lysosomal membranes. mTORC1 is recruited to the lysosome by RAPTOR and the RAG GTPases, and then activated in a parallel manner by RAG and Rheb GTPases. In this preprint, a model is described where Rheb exerts its activation effect on mTORC1 from its position on ER tubules. The activation of mTORC1 is governed by several environmental input signals, conveying information about amino acid levels, energy levels and growth factor presence.
Why I choose this preprint
As Rheb and mTORC1 are major regulators of cell growth and survival, these genes are found to be overexpressed in many forms of cancer1. Thus, it is important to understand the molecular regulation of Rheb localization and mTORC1 activation as this could provide information on how to interfere with this signaling in order to combat disease. While a lot of questions still remain, this preprint adds to our understanding of Rheb localization and describes how a very weak and transient interaction can be of functional importance.
I also appreciate that the authors of this preprint highlight the importance of critical analysis of immunofluorescence data, as fixation and preservation can influence antibody signal and structure preservation. Especially with broadly distributed proteins, false positive colocalization can often occur and it is of great importance to back up immunofluorescence data with other evidence.
Open questions
- What would be the explanation of the inability of Rheb to activate mTORC1, when constitutively localized to the ER?
- If the activation of mTORC1 is the result of transient interactions, why can these interactions not be obtained via cytosolic Rheb, or transient interaction between Rheb and the lysosome? What is the advantage of ER interactions?
- While the data in this preprint present evidence that Rheb localizes to the ER rather than the lysosome, there is a lack of discussion on how this reconciles with data showing Rheb localization to golgi6. Does farnesylation of Rheb mediate its interaction with both organelles? Furthermore, do the overlapping signals between lysosomes and ER correspond to contact sites between the organelles, and is this the sites of Rheb-mTORC1 interaction? Do these contact sites change upon refeeding, similar to what was reported for contact sites between lysosomes and Golgi?
- The localization pattern of GFP-Rheb looks rather distinct from the localization of endogenous Rheb. Is this simply due to a stronger signal of the GFP construct or could the overexpression of GFP-Rheb affect its localization and/or organellar structures? Overexpression of Rheb would presumably lead to a constitutively active mTORC1, which has been shown to induce ER stress7. Do subcellular fractionation of cells expressing GFP-Rheb show the same Rheb localization as WT cells?
- The model presented in this preprint does not mention the localization of the TSC1/TSC2 complex. Is it possible that this complex also resides on ER tubules to mediate its regulatory effect on Rheb, or is this regulation also facilitated by transient interactions between lysosomal TSC1/TSC2 and ER bound Rheb?
References
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012, 149(2): 274-293.
- Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem 2003, 278(35): 32493-32496.
- Kim SG, Buel GR, Blenis J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells 2013, 35(6): 463-473.
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem 2005, 137(3): 423-430.
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochemical and biophysical research communications 2006, 344(3): 869-880.
- Hao F, Kondo K, Itoh T, Ikari S, Nada S, Okada M, et al. Rheb localized on the Golgi membrane activates lysosome-localized mTORC1 at the Golgi-lysosome contact site. J Cell Sci 2018, 131(3).
- Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 2008, 29(5): 541-551.
doi: https://doi.org/10.1242/prelights.8185
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Deletion of PIEZO1 in adult cardiomyocytes accelerates cardiac aging and causes premature death
Theodora Stougiannou
Megakaryocytes assemble a three-dimensional cage of extracellular matrix that controls their maturation and anchoring to the vascular niche
Simon Cleary
Fis1 is required for the development of the dendritic mitochondrial network in pyramidal cortical neurons
Felipe Del Valle Batalla
preLists in the cell biology category:
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Jonathan Townson, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (2 votes)
(2 votes)