Mitochondrial biogenesis is transcriptionally repressed in lysosomal lipid storage diseases
Posted on: 6 March 2019 , updated on: 8 March 2019
Preprint posted on 6 February 2019
Putting the brakes on mitochondrial biogenesis: how lysosomal dysfunction can influence mitochondrial dynamics
Selected by Sandra Franco IborraCategories: cell biology
Mitochondria evolved from a bacterial ancestor which, through the symbiosis with an eukaryotic host cell (Margulis, 1970), lost nearly all of its functions as a separate entity over the course of time and became the mitochondrion. However, mitochondria still maintain their own genome, coding for 13 proteins in mammalian cells. Therefore, the vast majority of the mitochondrial proteome is encoded in the nucleus, synthesized in the cytosol and imported into the organelle (Wiedemann et al 2004). As a result, the generation of new mitochondria, also called mitochondrial biogenesis, requires a complex coordination of the nuclear and mitochondrial expression programs. This is achieved by specific nuclear transcription factors that regulate the expression of nuclear genes encoding mitochondrial proteins, such as Nuclear Respiratory Factor 1 (NRF1). Besides transcription factors, there are also co-activators, such as peroxisome proliferator-activated receptor-gamma co-activator 1 α (PGC1α), which also participate as positive regulators in mitochondrial biogenesis (Scarpulla et al 2012). Due to the central role that mitochondria play in cellular homeostasis, it comes as no surprise that alterations in mitochondrial functions are present in a wide variety of diseases.
In this preprint, the authors have taken a close look at mitochondrial dysfunction in two lysosomal storage diseases: Niemann-Pick type C (NPC), caused by mutations in genes that encode proteins involved in efflux of lipids from the lysosome; and shingomyelinase (ASM) deficiency, caused by mutations in the gene encoding ASM, which catalyzes the degradation of sphingomyelin.
Key findings
The authors uncovered specific down-regulation of mitochondrial transcripts in both NPC and ASM deficiency, pointing at altered mitochondrial homeostasis as a common mechanism in lysosomal storage disorders. Despite the transcriptional repression, mitochondrial mass was increased in fibroblasts from NPC and ASM patients. This can be explained by the lysosomal loss of function present in lysosomal storage disorders, which leads to accumulation of cargoes normally degraded by this organelle, such as mitochondria. Mitochondrial dysfunction, characterized by increased ROS production and decreased mitochondrial respiratory capacity, was also present in patient’s fibroblasts, probably due to the accumulation of dysfunctional mitochondria.
Next, the authors wondered which transcriptional regulators could mediate the repression of mitochondrial-associated genes. Upon analysis of the promoters of those mitochondrial genes whose expression was repressed, they found two transcription factor families, Krueppel-like factors (KLF) and ETS factors that were enriched. Specifically, KLF2 and ETV1 transcription factors showed increased activity in brain and liver from NPC KO mouse. To further determine whether KLF2 and ETV1 were repressing mitochondrial biogenesis, the authors knocked down both genes independently in ASM-deficient fibroblasts, which rescued the transcript levels of mitochondrial proteins that were downregulated, including important mitochondrial biogenesis regulators such as NRF1, its downstream target TFAM, and NRF2. Although silencing of KLF2 and ETV1 partially rescued mitochondrial respiratory capacity, lysosomal function remained impaired.
The next question to tackle was the relationship between KLF2 and ETV1. The authors found that KLF2 is located upstream of ETV1 and can modulate ETV1 expression. Moreover, Erk signaling is necessary for KLF2-dependent activation of ETV1. And what about KLF2 regulation? KLF2 turns out to be part of a signaling network of sphingosine-1-phosphate receptor 1 (S1PR1), by which KLF2-mediated transcriptional activation of S1PR1 acts as negative regulator of KLF2 activity. Interestingly, inhibition of S1PR1 leads to increased KLF2 expression, repression of mitochondrial proteins and altered mitochondrial oxygen consumption rate. However, when treating ASM-deficient fibroblasts with an S1PR1 agonist no changes were observed. Indeed, ASM-deficient fibroblasts display decreased levels of S1PR1 receptor in the plasma membrane without any changes at the protein level, suggesting that S1PR1 mislocalization leads to increased KLF2 signaling and its downstream consequences. Importantly, KLF2 and ETV1 upregulation represents a protective mechanism in ASM-deficient fibroblasts, since the silencing of these two mitochondrial biogenesis repressors leads to increased cell death.
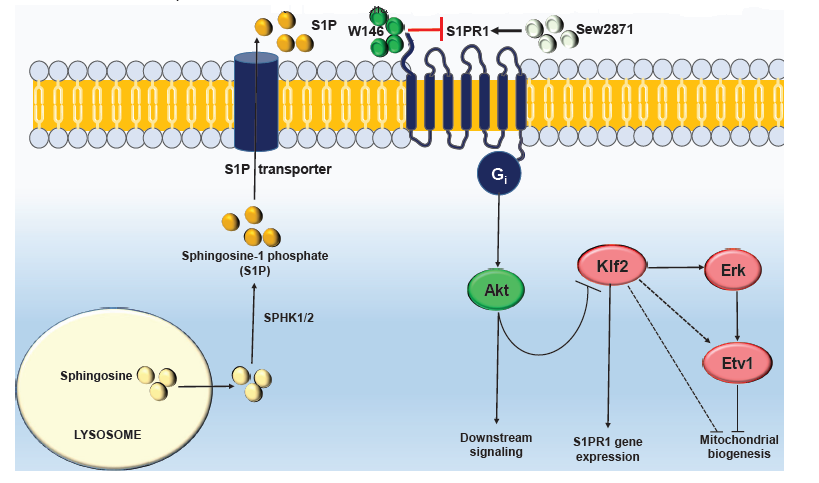
Schematic illustration of sphingosine-1-phosphate (S1P) signaling. S1P is synthesized inside the cell and transported to the extracellular space through S1P transporter. Extracellular S1P activates sphingosine-1-phosphate receptor 1 (S1PR1), which triggers Akt downstream signaling, including KLF2 repression. Upon loss of S1PR1 receptor and/or S1P signaling, KLF2 is activated and can induce its downstream targets, leading to repression of mitochondrial biogenesis.
Why I like this preprint
In the field of mitochondrial biology, there is this assumption that mitochondrial biogenesis relies on a subset of transcriptional factors and coactivators that positively regulate the mitochondrial transcriptional program. But less is known about repressors of mitochondrial biogenesis. This preprint sheds light on the existence of mitochondrial transcriptional “brakes” which could also play a role in mitochondrial disorders and neurodegenerative diseases characterized by mitochondrial dysfunction. Another key aspect of this preprint is the underlying idea of the importance of organelle crosstalk in health and disease. This organelle communication can take place through membrane-membrane contacts or involve complex cellular signaling. In any case, it is clear that alterations in any component of the network can have important consequences for other elements, as it is illustrated in this preprint showing that lysosomal dysfunction gives rise to mitochondrial biogenesis repression.
Questions for the authors
- The authors showed how in conditions where lysosomal function is impaired and mitochondria are not being degraded, there is a shutdown of mitochondrial biogenesis. This repression seems to be orchestrated by the internalization of S1PR1 and decreased sphingosine-1-phosphate signaling, which in turns activates KLF-2 and its downstream transcriptional program. Which mechanism is responsive for S1PR1 internalization?
- Silencing of KLF2 and ETV increases mitochondrial biogenesis and partially normalizes the mitochondrial respiratory capacity in ASM-deficient fibroblasts. However, no improvements are observed regarding lysosomal function. Is the increase of mitochondrial biogenesis compensating for the damaged undegraded mitochondria accumulated in the cytosol? At the same time, silencing of KLF2 and ETV1 in ASM-deficient fibroblasts leads to increased cell death, pointing at KLF2 and ETV1 activation as a protective mechanism in ASM deficiency. Does this imply that the mitochondrial dysfunction observed in lysosomal storage disorders minimally contributes to the disease.
References
- Margulis, L. (1970). Origin of eukaryotic cells. New Haven (Connecticut): Yale University Press.
- Wiedemann N. et al. The protein import machinery of mitochondria. J. Biol. Chem. 279, 14473–6 (2004)
- Scarpulla R. et al. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol. Metab. 23, 459-469 (2012).
doi: https://doi.org/10.1242/prelights.9127
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)