A genome-wide knock-out screen for actors of epigenetic silencing reveals new regulators of germline genes and 2-cell like cell state
Posted on: 15 June 2021
Preprint posted on 3 May 2021
Article now published in Nature Structural & Molecular Biology at http://dx.doi.org/10.1038/s41594-023-01038-z
A Dazling approach: CRISPR KO screen sheds some (red) light into the missing pieces of the epigenetic puzzle in mESC
Selected by María Mariner-FaulíCategories: developmental biology, genetics, genomics
Background
At a time when increasing amounts of omics data are being generated on a daily basis, it is becoming even more important to functionally characterise genes in a context-dependent manner. In this regard, the application of genome-wide CRISPR-Cas9 screenings in different cell culture models emerge as powerful large-scale approaches to identify promising candidates whose disruption elicits a specific phenotype or physiological effect.
The rationale behind this technique starts by choosing the specific readout that cells will manifest after the screening (i.e drug resistance, cell reprogramming) and knocking out every gene that could be implicated in this readout. Importantly, only one gene per cell will be knocked-out, and after incubation, some cells will start to show an altered phenotype, providing a list of candidate genes that can be further studied.
Genome-wide CRISPR-Cas9 screen can be designed for a wide variety of applications. And here is where the fun part begins! In order to identify key regulators of developmental transitions, different reporter systems have been engineered as optimal readouts of fate transition [1,2,4]. This preprint by Nikhil Gupta and colleagues is an example of the great utility of these reporter systems in conjunction with whole genome screening, and provides an interesting list of novel regulators of early pluripotency states and germline genes.
Summary
Epigenetic mechanisms represent an important regulatory layer essential both for the maintenance of cellular identities and for the optimal transition between different developmental states. The main actors behind these regulatory processes are the commonly named epigenetic “writers” and “erasers”: different factors that establish or remove epigenetic marks and whose modifications will be interpreted by several “readers”, translating them into a transcriptional response. Thanks to massive screening techniques, the catalogue of epigenetic effectors is rapidly increasing, providing a progressively more complete picture of the complexity of epigenetic regulatory mechanisms.
In this preprint, Nikhil Gupta and colleagues establish a mESC-based reporter system responsive to epigenetic modifications. In a search for repressors, they take advantage of the tunability of the epigenetic state of mESCs depending on culture conditions. On the one hand, cells cultured in 2i+LIF are grown in the presence of two inhibitors of MEK/ERK and GSK3b signalling pathways plus leukemia inhibitory factor. As a consequence, they have lower DNA methylation levels and show features of naïve pluripotency. On the other hand, cells in S+L are cultured in the presence of fetal bovine serum (undefined chemical composition) and leukemia inhibitory factor, and show a repressive and methylated chromatin landscape and display features of metastable pluripotency.
In this specific transition context, it becomes crucial to choose an optimal endogenous reporter gene. In the search for the perfect candidate, the authors considered three main criteria: epigenetic mechanisms of repression, biological function of the gene and its expression level. Taking all three into consideration, they chose Dazl (a germ cell and 2C developmental stage marker) as a reporter gene given its expression (2i=ON, S/L= OFF) and methylation pattern (2i=unmethylated, S/ L=methylated). Next, they engineered a cell line with a monoallelic knock-in of two selection markers (mScarlett and hygromycin resistance) driven by the Dazl promoter (DASH cell line). Interestingly, Dazl is informative as a reporter for genome wide CRISPR-Cas9 screens, as its repression depends on the synergistic action of ncPRC1.6 and DNA methylation, broadening the search for putative repressors. This DASH reporter cell line is the key tool of the study, and is subjected to a CRISPR-Cas9 genome-wide screening with the Brie library, a lentiviral library of vectors co-expressing sgRNAs and Cas9, targeting approximately 20000 genes [3].Following this experimental pipeline, the authors reach the goal of the study: the identification of repressive factors whose knock out triggers gene reactivation in serum condition (Figure 1).
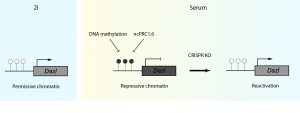
Figure 1: Graphical representation of the reporter system used in the study. In 2i conditions, Dazl is expressed while in S/L it is repressed trough DNA methylation and ncPRC1.6 complex. Knocking out factors essential for repression either at local or global scale triggers the reporter reactivation in serum condition. CpG DNA methylation status is represented by white (unmethylated) or black (methylated) lollipops. Figure modified from Gupta et al. 2021 (preprint).
The screening yielded a list of expected hits (DNMT1, UHRF1, MGA) involved in DNA methylation maintenance. These results served as an initial validation of the reporter system After narrowing down the list of candidates based on its statistical significance and biological function, the authors focused on six genes that were not known to regulate Dazl: Zbtb14, Kdm5c, Spop, Mcm3ap, Bend3 and Kmt2d. Next, they performed independent knock outs of these candidates in the DASH cell line in order to validate the results initially obtained in heterogeneous KO populations. Interestingly, the knock-outs of Kmt2d and Kdm5c resulted in the highest Dazl inductions. They confirmed that the observed effects were strictly due to the knock outs by rescue experiments, which reduced the mScarlet signal and sensitized cells to hygromycin. Then, they proceeded to further characterize the knock-outs by transcriptomics and methylation analysis. PCA analysis based on the transcriptome of the knock-outs grouped together Kdm5c, Mcm3ap, Spop and Zbtb14, while Kmt2d and Bend3 remained isolated, close to serum grown cells. Regarding methylation levels, only Spop KO showed global hypomethylation in comparison to parental cells.
In addition, knock out of Zbtb14, Kdm5c, Spop and Mcm3ap reactivates the poorly characterized 2-cell like cell (2CLC) state, an in-vitro model of totipotency, whose transcriptome highly overlaps with that of two-cell stage embryos and models the transition from maternal to zygotic expression program in early mouse development
Altogether, this work describes an endogenous DNA methylation-sensitive reporter system that can be useful in the search for transcriptional repressors in different cell state transition models and using different representative loci as reporters. Last but not least, the work by Defoussez´s Lab opens the door for further characterization of the novel epigenetic repressors discovered in this study.
Key results
- Development of DASH reporter cell line as a suitable reporter system for the study of epigenetic repressive pathways.
- Discovery of novel epigenetic repressors of germline genes, repeat elements and 2-cell like cell state, prioritizing ZBTB14, KDM5C, SPOP, MCM3AP, BEND3 and KMT2D.
Why I chose this preprint
As part of my daily twitter scroll early in May I found this preprint from Defossez´s Lab about genome-wide CRISPR KO screening. Turns out I had been recently hanging out with a friend who is applying it to cancer research and we had been discussing the technique´s potential as a massive search for oncogenes. Therefore, I decided to proceed to further reading, finding a very interesting experimental approach that not only helped me gain some theoretical background, but also gave me new ideas for my project. The perfect Prelight, isn´t it?
Epigenetic mechanisms represent a complex and context-specific set of regulatory processes whose coordination rules are still far from being fully understood. The regulatory and signalling cascades dictating cell identity and cell fate transitions still miss molecular effectors in order to get global pictures of the mechanisms involved. These new discoveries are of crucial relevance for the prevention, diagnosis and treatment of several developmental syndromes and diseases such as cancer.
This preprint adds to recent works searching for epigenetic regulators using endogenous reporters whose expression is dependent on effectors involved in the regulatory mechanisms explored [1,4]. These publications are providing the missing pieces of the epigenetics puzzle, and further study will provide more accurate predictions on the mechanisms ruling cell identity.
Questions for the authors
- The authors mention that one limitation of this reporter approach is the loss of information from the knock out of essential or near-essential genes. Another possible restraint of the study could be the presence of false negatives caused by mechanisms of redundancy between transcription factors and/or transcriptional compensation due to the absence of full gene knock outs. How could the authors overcome such limitations in further studies?
- What is the rationale behind using two selection markers (mScarlet and Hygromycin resistance) rather than using only mScarlett and filtering positive cells by FACS?
- One of the top hits is KMT2D (MLL4), which is considered as an activator mostly in the context of enhancers. How can KMT2D contribute to Dazl repression? Could the repression of Dazl be an indirect effect due to silencing of other genes in the absence of KMT2D? How can the authors distinguish between direct and indirect regulators of Dazl?
References
- Dixon, et al. QSER1 protects DNA methylation valleys from de novo methylation. Science 372, (2021).
- Alda-Catalinas, C. et al. A Single-Cell Transcriptomics CRISPR-Activation Screen Identifies Epigeneti- Regulators of the Zygotic Genome Activation Cell Systems 11, 25-41.e9 (2020).
- Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPRCas9. Nature Biotechnology 34, 184–191 (2016).
- Douillet, D., Sze, C., Ryan, C. et al. Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nature Genetics 52, 615–625 (2020).
doi: https://doi.org/10.1242/prelights.29562
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Dosage-sensitive RBFOX2 autoregulation promotes cardiomyocyte differentiation by maturing the transcriptome
Theodora Stougiannou
Post-translational Tuning of Human Cortical Progenitor Neuronal Output
Jawdat Sandakly
Also in the genetics category:
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
VANGL2 shapes the mouse heart tube from adjacent epithelia and without planar polarity
Anubhav Prakash
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genetics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)