Imaging cell lineage with a synthetic digital recording system
Posted on: 16 April 2020
Preprint posted on 25 February 2020
Article now published in Science at http://dx.doi.org/10.1126/science.abb3099
Imaging of cells recording their own lineage history with intMEMOIR, or why lineage research is more alive than ever
Selected by Irepan Salvador-MartinezCategories: developmental biology, genetics
Introduction
Cell lineage research is experiencing a renaissance. Already in the late 19th century researchers were interested in describing and comparing cell lineages of different organisms (Wilson, 1898). For many decades, however, the investigation of cell lineages was limited by the lack of techniques that could be used to track the cell division of complex organisms. That has changed now. In recent years, newly developed technologies promise to finally allow the reconstruction of the lineage history of a complex organism.
A popular approach in recent years has been to use CRISPR-based editing systems that generate heritable mutations during the development of an organism (e.g. McKenna et al, 2016). The mutations inside the cells of such organisms are usually recovered by sequencing, which leads to the loss of the spatial context of the cells. A major goal of current research is, therefore, to describe the lineage history of the cells together with their identities (cell types) and spatial distribution.
About the preprint
Chow, Budde and collaborators designed a new lineage recording system (intMEMOIR) that introduces mutations using serine integrases and use Fluorescence in situ Hybridisation (FISH) to read these mutations. Serine Integrases are bacteriophage enzymes that introduce mutations via site- specific recombination after they recognise and bind to attB and attPs sites.
The lineage recorder designed by the authors contains multiple recording units, each of which consists of a barcode sequence flanked by att sites (Fig 1). This configuration leads to either an inversion or a deletion of the barcode sequence after binding of the integrase. Therefore, each recording unit will end up in 1 of 3 possible states: “unmutated”, “inverted” or “deleted”. After a site is “inverted” or “deleted” it cannot further change and it’s inherited in subsequent cell divisions.
Importantly, each recording unit has a unique att site that prevents recombination between units. By linking each of these 3-state sites —or “trits”— to a strong promoter they could express 1) the forward barcode (“unmutated” state), 2) the reverse complement barcode (“inverted”) or 3) no transcript (“deleted”). These “trits” represent a considerable improvement from a previous approach that used 2-state units —or “bits”— (Frieda et al, 2016). This is because recorders made of 2-state units would get to the same end-state if they become completely saturated with mutations, making them uninformative.
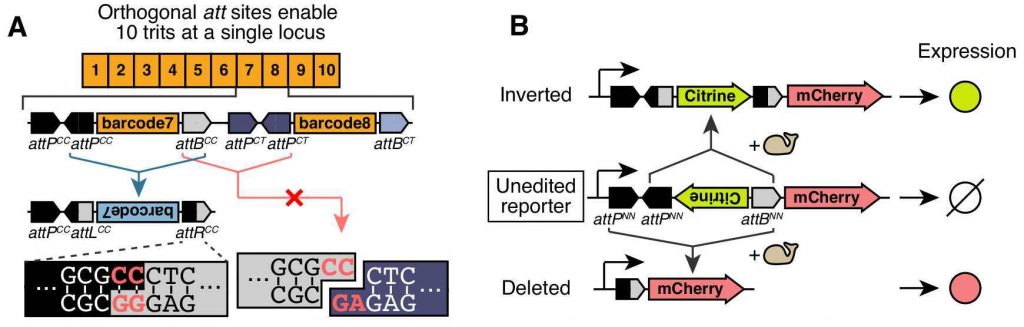
attP/attB pairs with orthogonal central dinucleotides (red letters). (B) Fluorescent reporter assay
enables rapid characterization of the recording units (from Figure 2 in the preprint made available under a CC-BY-NC-ND 4.0 license).
Results
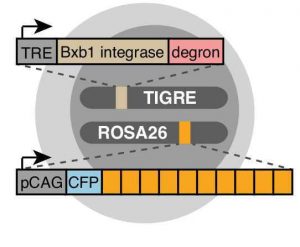
After validating the trit design in mouse embryonic stem cells, the authors engineered a cell line (intMEM1) with a recorder array of 10 trits (Fig 2) and where the recording activity can be activated in a controlled manner by the addition of doxycycline and trimethoprim (TMP) to the cell culture. The authors then devised a clever approach to decode the recorders’ mutations by five rounds of hybridisation chain reaction smFISH, using 4 fluorophores and 20 unique probes. Essentially, this approach leads to cells emitting up to 4 colours per round of hybridisation depending on the state of its lineage recorder and on the orientation of the transcribed barcode. The colours/states of all cells in the culture are then used to reconstruct their lineage relationship.
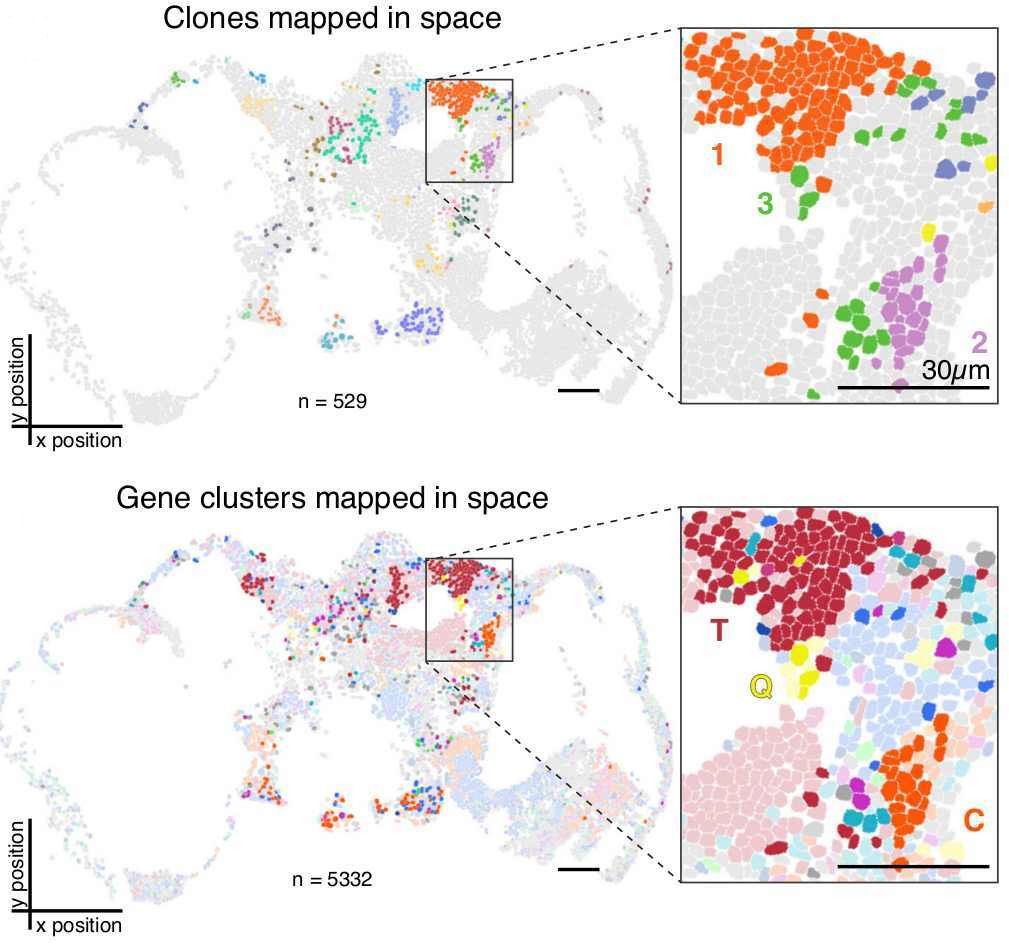
In order to asses the accuracy of reconstructed lineages, the authors compared these to ground-truth lineages obtained after tracking cell divisions on time-lapse movies and also made use of computer simulations. The ground truth lineages were based on cultures that underwent recording activity for ~3 generations and ~1-2 generations of clonal expansion. They assessed lineage reconstruction at two different levels: “clonal classification” and “tree reconstruction”.
For “clonal classification” cells that share the same exact barcode were grouped into “clones” and the agreement between these “clones” with the ground truth lineage were assessed. For “tree reconstruction”, the relationship between “clones” was inferred by reconstructing a lineage tree using a maximum likelihood approach incorporating empirical parameters. The accuracy of this tree was determined as the fraction of splits that is shared between the reconstructed and the ground truth trees (Robinson-Foulds metric). Importantly, these two levels of reconstruction (clones vs tree) can be used either together or individually, depending on the biological question. As the authors also show, clonal classification can be useful to analyse the clones’ spatial distribution in a tissue/organ without the necessity of reconstructing a completely accurate lineage tree – a task which remains difficult (Salvador-Martinez, et al 2019).
Their results showed a high accuracy of reconstruction at both levels, with an average of 85% of the cells inside a reconstructed clone being correctly assigned to it (“clone classification”) and 25% of clonal relationships reconstructed perfectly, with the overall reconstruction score significantly higher than the random control (“tree reconstruction”).
Finally, to test their recorder in vivo, they created a Drosophila melanogaster line (termed “Drosophila memoiphila”) with the intMEMOIR construct, where the integrase activity can be regulated via a heat-shock inducible promoter. They used this line to investigate clonal relationships in the fly brain. For this, they induced mutations during early development by heat shock, letting the flies develop until adults. Mutated adult flies were then collected and their brain sectioned for smFISH imaging. As the same imaging approach could be used to read transcripts from endogenous genes, the authors decided to read, in top of the lineage recorder units, 8 genes that could discern between different neuronal cell-types. The combination of gene expression, lineage information, and spatial location within the brain allowed them to identify different known cell types (e.g. GABAergic and dopaminergic neurons) and interrogate how their cell type correlated with their cell lineage. They found that cells of the same clone showed stronger cell type similarity to each other the more physically closer they were. Importantly, this relationship between physical distance and cell type similarity was not observed when comparing cells of different clones.
Why I chose this preprint
With this exciting novel approach, Chow, Budde, and collaborators remind us again about the importance of knowing the lineage history of the cells in order to have a better understanding of their differentiation within their native spatial context, and introduce a new fly line “Drosophila memoiphila” than can be used as a general purpose lineage analysis resource. By doing so, they prove that cell lineage research is alive, rejuvenated and has a promising future.
Question to the Authors:
Q1: One exciting possibility that you suggest is to increase the lineage information that can be read by the incorporation of additional arrays. Is there a limit on the number of FISH readout cycles that can be performed on a slide?
Q2: I find very interesting how the Shannon’s entropy of a mutated array relates to its accuracy of reconstruction. Could the Shannon’s entropy measure serve to determine a reliability threshold for mutated arrays?
Q3: How easy is to differentiate between a “deleted” state from the possibility of having another state but a lack of signal?
References
Wilson, E.B. CONSIDERATIONS ON CELL‐LINEAGE AND ANCESTRAL REMINISCENCE. Annals of the New York Academy of Sciences, 11: 1-27 (1898). doi:10.1111/j.1749-6632.1898.tb54960.x
McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science (2016). https://doi.org/10.1126/science.aaf7907
Frieda KL, Linton JM, Hormoz S, et al. Synthetic recording and in situ readout of lineage information in single cells. Nature. (2016). https://doi.org/10.1038/nature20777
Salvador-Martínez I, Grillo M, Averof M, Telford MJ. Is it possible to reconstruct an accurate cell lineage using CRISPR recorders? eLife. 2019;8. https://doi.org/10.7554/eLife.40292.001
doi: https://doi.org/10.1242/prelights.18724
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cross Sectional and Longitudinal Imaging Reveals Spatiotemporal Divergence in Morphogenesis and Cell Lineage Specification between in-vivo and in-vitro Mouse Embryo during Pre- and Peri-implantation
Heather Pollington
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Also in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |











 (No Ratings Yet)
(No Ratings Yet)