Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA clusters in cancer
Preprint posted on 21 October 2020 https://www.biorxiv.org/content/10.1101/2020.10.20.335216v1
Categories: cancer biology, cell biology, molecular biology, neuroscience
Background1-3
Extrachromosomal DNAs (ecDNAs) are genetic elements present inside the nucleus independent of a linear chromosome that are about 50kb-5Mb in size, harboring a set of genes and regulatory elements. ecDNAs are formed when broken linear DNA ligate end-to-end. ecDNAs are found in many eukaryotic species like yeast, D. melanogaster, C. elegans, and humans. In cancer cells, ecDNAs predominantly accommodate oncogenes or fused oncogenes to facilitate oncogene amplification. However, it is not clear how ecDNA propagate intratumor genetic heterogeneity, a characteristic feature of multidrug-resistant cancer colonies. Unequivocal segregation of ecDNA to daughter cells could impel intratumor genetic heterogeneity. To address this, the authors of the current study used cutting-edge CRISPR based imaging techniques to investigate the dynamics of ecDNA in glioblastoma.
Key findings
1. To gain insights into the dynamics of ecDNA in glioblastoma, the investigators used four glioblastoma samples and a pair of neurospheres derived from the same patients. They evaluated fluorescent in situ hybridization (FISH) of EGFR(an oncogene of glioblastoma) to score ecDNA based on their earlier studies4 and a chromosome 7 (Chr7) loci for linear intact chromosome. They found that EGFR-containing ecDNA numbers vary among samples than Chr7 that was rather evenly distributed. They further report that unequivocal EGFR ecDNA amplification corroborated with heterogeneity in protein expression. In other cell lines, they also found heterogenous gene amplification of ecDNA-associated genes than genes harbored in linear chromosomes. Thus, they report that ecDNA could drive genetic heterogeneity in cancer cells (fig.1).
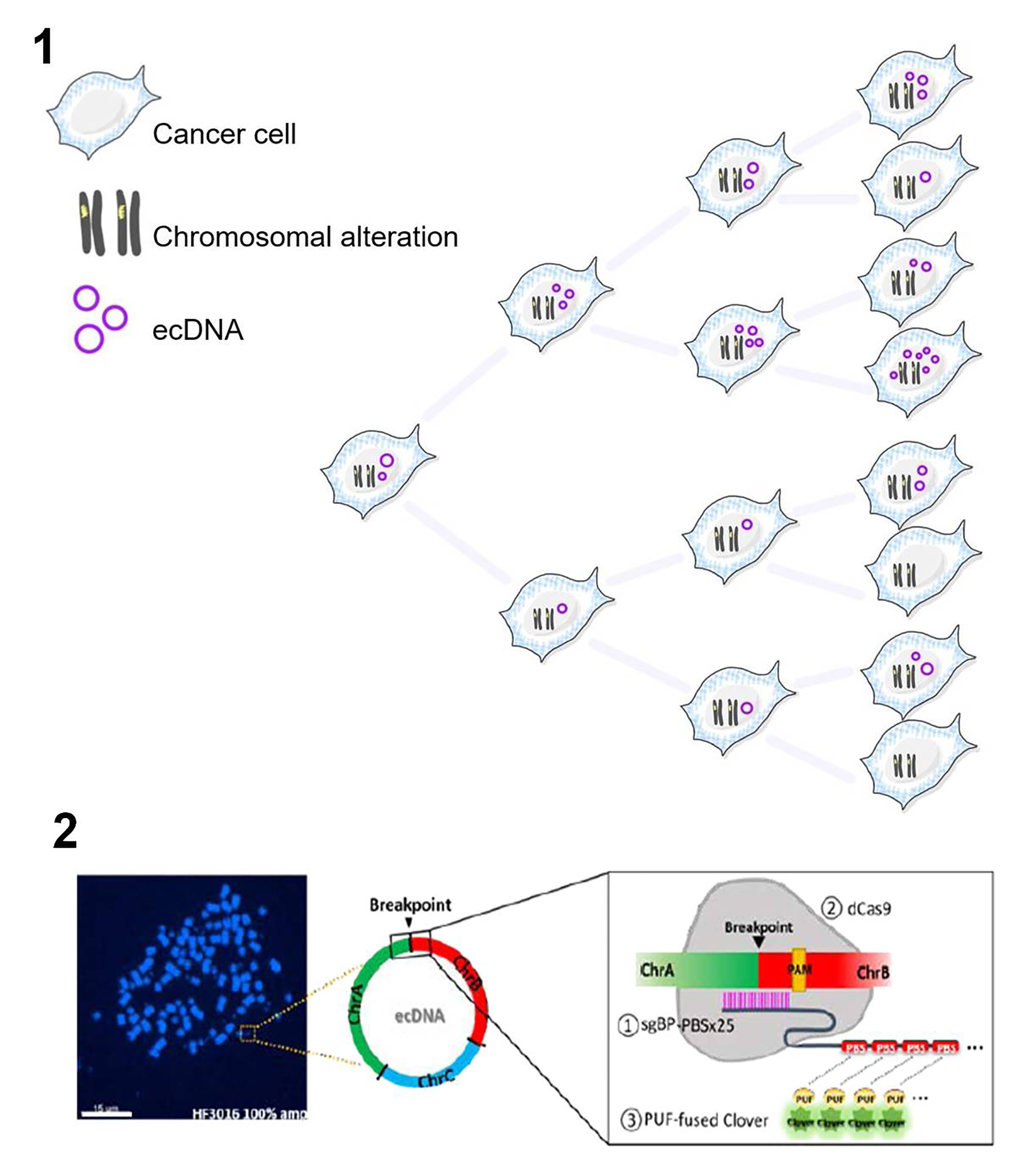
2. To address how ecDNA heterogeneity is dispersed in cancer cells, the authors implemented a CRISPR-based DNA labeling system (fig.2). For this purpose, they used nuclease dead Cas9, provided a single guide-RNAs with 25 Pumilio/FBF (PUF) RNA-binding sites that would bind to the unique fusion sequences at the ecDNA breakpoints, and recruit fluorescently tagged PUF proteins to identify ecDNA in real-time. They identified four unique ecDNAs – ecEGFRx1, ecEGFR, ecCCAT1, and ecCCDC26– harboring an EGFR exon1, a full-length EGFR, and non-coding genes CCAT and CCDC26(using sequencing, FISH, and PCR-based techniques). They detected all four ecDNA outside chromosomes using breakpoint-specific FISH imaging in metaphase spreads. They were able to demonstrate all four ecDNA using the CRISPR-based system in neurosphere cells but not in prostate cancer cells that acted as a control. Moreover, they found that CRISPR labeling coincided with the breakpoint-specific FISH signal suggesting the reliability of the signal. They also found heterogeneous ecDNA copy numbers than linear chromosome locales (Chr7 and MUC4) in the neurospheres.
3. The authors then tracked the ecDNA live using the same CRISPR-based DNA labeling system and found uneven segregation of the ecDNA to daughter cells even when followed for 48hrs, unlike Chr7and MUC4. They also found that ecDNAs tend to coalesce in at least 50% of neurospheres in the 48hr time frame. Furthermore, they show that these ecDNA clusters co-localized with nuclear bodies5 (Cajal and PML bodies) that act as a macromolecular hub, but there was no significant linear correlation between the number of ecDNAs and the number of nuclear bodies, suggesting that ecDNAs generate its own hub. Intriguingly, they also report ecDNA clusters co-localized with RNA polymerase II in 60% of the cells and the size of ecEGFRfoci indicating its clustering positively correlated with EGFR mRNA levels.
Conclusion
Recent studies revealed intratumor genetic heterogeneity as a critical factor for the perpetuation of multidrug-resistant cancer colonies. Intratumor genetic heterogeneity poses a great challenge to personalized medicine. Here the authors report that intratumor genetic heterogeneity is promoted by ecDNAs through their uneven segregation during cell divisions and clustering to facilitate oncogene expression or amplification (fig.1). The current study raises many interesting hypotheses on how genome instability drives oncogenesis and cancer evolution.
Acknowledgments
I am grateful to all the authors for their support, especially Eunhee Yi and Roel GW Verhaak for being open to discuss the work and replying promptly.
References
- https://doi.org/10.1038/s41568-019-0128-6
- https://doi.org/10.1016/j.molonc.2014.06.005
- https://doi.org/10.1016/j.annonc.2020.03.303
- https://doi.org/10.1038/s41588-018-0105-0
- https://doi.org/10.1016/S0962-8924(99)01606-2
Posted on: 28 October 2020
doi: https://doi.org/10.1242/prelights.25485
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cancer biology category:
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Invasion of glioma cells through confined space requires membrane tension regulation and mechano-electrical coupling via Plexin-B2
Modeling transcriptional profiles of gene perturbation with a deep neural network
Also in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Also in the molecular biology category:
Fetal brain response to maternal inflammation requires microglia
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
Also in the neuroscience category:
Fetal brain response to maternal inflammation requires microglia
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
preLists in the cancer biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the neuroscience category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)