YAP1 Regulates the Self-organized Fate Patterning of hESCs-Derived Gastruloids
Posted on: 25 June 2021
Preprint posted on 12 March 2021
YAP1 transcriptionally represses NODAL genes to ensure appropriate positioning and size control of the 3 germ layers in human gastruloids.
Selected by Srivatsava ViswanadhaCategories: biophysics, cell biology, developmental biology
Background:
Gastrulation is the morphogenetic event that gives rise to the three founder lineages of all organs and tissues – ectoderm, mesoderm, and endoderm – from the pluripotent epiblast. Beyond generating these three germ-layers, gastrulation must ensure their proper location and adequate size. This is achieved through the spatio-temporal regulation of the Nodal pathway [1,2]. While Nodal inhibition prompts acquisition of neuroectoderm potential, its activation with cooperative action from BMP and Wnt signaling directs the formation of the other two lineages [3,4].
Nodal antagonists are secreted by the non-pluripotent cell population in mice and human blastocysts [5,6]. However, the spatial patterning of all three germ layers has been recapitulated by solely culturing Embryonic Stem Cells (ESCs), which are the in vitro counterparts of the pluripotent epiblast. When cultured in defined media conditions, human ESCs (hESCs) undergo morphological and molecular changes reminiscent of gastrulation. Hence, they are referred to as “gastruloids”. Importantly, fate patterning in both 2D hESC cultures and symmetry breaking in 3D gastruloids were achieved without the precise spatial regulation of the Nodal pathway [7,8]. This hints at the existence of a hitherto unknown cell-intrinsic mechanism which regulates Nodal spatial distribution and thereby governs lineage size and positioning.
In this preprint, the authors elucidate the molecular logic behind Nodal-mediated fate patterning. They show that the size and location of germ layers and fate choice of hESCs are dictated by the transcriptional regulator YAP1. To achieve this, they employ a robust 2D model of human gastrulation based on micro-patterned hESCs. The key experimental techniques performed include directed differentiation, immunofluorescence, confocal microscopy, western blotting, and genome sequencing analysis.
Key findings:
-
YAP1 depletion alters the size and location of 3 discrete germ layers
Previous work [9] identified the fate restriction role of YAP1 during hESC differentiation. The rationale for focusing on YAP1 also includes its effector function in the Hippo pathway for organ size control. Therefore, the authors surveyed the gastrulation outcome of YAP1 knock-out cells (YAP1-KO). To do so, they confocally imaged hESC 2D-gastruloids immunostained for the putative lineage markers of the 3 germ layers SOX2 (ectoderm), SOX17 (endoderm), and Brachyury/T(Mesoderm). Through image analysis, they observed that YAP1-KO samples had a reduction in the share of SOX2+ve ectodermal cells and a compensatory increase in the individual proportions of mesodermal and endodermal cells. Moreover, the organization of the three germ layers was disrupted: (i) ectoderm located in the center of wild-type (WT) gastruloids versus at the colony periphery in YAP1-KO gastruloids and (ii) thicker mesendoderm in YAP1-KO gastruloids (Figure.1 A&B).
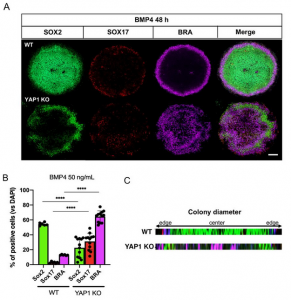
-
YAP1 promotes ectodermal specification by cytoplasmic retention of SMAD-2.3
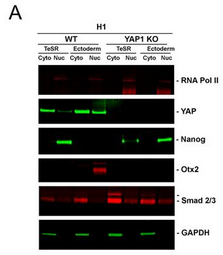
Since the percentage of ectodermal cells was reduced upon YAP1 depletion, the authors wanted to test if this was due to an inherent inability of fate decision making. For this, they performed directed differentiation towards individual germ layers. Through transcriptomic analysis, they observed that despite biochemical ectoderm induction, YAP1-KO hESCs continued to express high levels of core pluripotency factors including Pou5f1 (Oct4) and specifically Nanog. The latter is known to repress neuronal differentiation through its functional antagonism of Otx2 [10], an ectoderm specifier. By rescuing YAP1 levels through an inducible expression system, the authors were able to restore Nanog downregulation and Otx2 expression.
Having confirmed the repression of Nanog, the authors wanted to identify the molecular framework for YAP1’s inhibitory action. For this, they focused on investigating changes in the levels of SMAD-2.3 and SMAD-1.5 in WT vs YAP1-KO cells. SMAD-2.3 and SMAD-1.5 are respectively the effectors of NODAL and BMP signaling pathways, the two cascades which inhibit ectoderm fate acquisition. More importantly, nuclear SMAD-2.3 drives Nanog expression [11], the phenotypic outcome of YAP1-KO cells in directed differentiation experiment. Therefore, the authors performed western blotting with antibodies targeting the aforementioned SMADs. Additionally, the protein lysates were separated into nuclear and cytoplasmic fractions to observe changes in both overall protein level and subcellular distribution. While SMAD-1.5 had no discernible changes, SMAD-2.3 levels increased in YAP1-KO cells, and it was predominantly localized in the nucleus. The authors tested the relevance of this result through neuronal induction of geometrically confined hESCs. In line with earlier observations, YAP1-KO cells exhibited predominantly nuclear SMAD-2.3 and lacked the expression of the ectodermal marker Sox2 (Figure.2).
Thus, the authors proved that ectodermal specification requires YAP1-mediated inhibition of nuclear SMAD-2.3 activity.
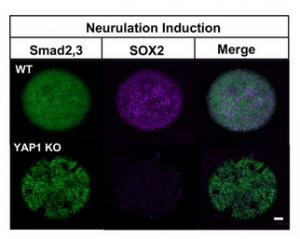
-
Direct transcriptional regulation of developmentally relevant genes by YAP1
The high nuclear levels of SMAD-2.3, a downstream component of the Nodal cascade, prompted the authors to test for expression levels of the Nodal signaling pathway in ectoderm-induced WT and YAP1-KO cells. For this, they utilized the RNA-seq datasets obtained from directed differentiation experiments and performed Signal Pathway Analysis. After filtering the results based on relevance and statistical significance, the authors observed that YAP1-KO hESCs were enriched in genes driven by Nodal signaling. When RT-qPCR was performed for the selected NODAL genes after inducible YAP1 rescue, their levels were found to be reduced. This confirms the transcriptional repression action of YAP1 on NODAL pathway genes.
To gain further insight into the regulatory mechanism of YAP1, the authors performed single-nuclei ATAC sequencing. This technique identifies transcriptionally accessible chromatin regions of the genome. The differentially expressed regions can be categorized into proximal and distal which rely on promoter-based and enhancer-based transcriptional regulation, respectively. When the authors probed the loci that gained transcriptional accessibility upon YAP1 depletion, these were found to be upstream of NODAL and FOXH1 genes. ChiP-Seq analysis did not only confirm but also identify those putative-YAP1-binding sites. Importantly, the YAP1-bound region of NODAL was found to be the well-characterized Proximal Epiblast Enhancer (PEE). Further analysis identified a conserved YAP1 binding site of PEE in the mouse genome. This suggests an evolutionarily conserved role of YAP1 for NODAL regulation during gastrulation.
Finally, the authors wanted to confirm if the observed gastrulation defects in YAP1-KO hESCs were due to upregulated NODAL signaling. For this, they performed a 2D gastrulation assay using YAP1-KO with NODAL inhibition. With this approach, they were able to rescue the fate patterning defect. Ectoderm was restricted to the center and the other two lineages to the periphery of the colony. Moreover, directed differentiation proved that NODAL inhibition rescued the ability of YAP1-KO cells to be specified towards the ectodermal lineage (Figure.3).
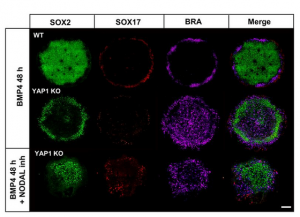
Conclusion:
This work proves that YAP1 ensures appropriate fate patterning during hESC gastrulation via its transcriptional repression of NODAL signaling. The transcriptional regulation is expected to be mediated through YAP1 binding to the NODAL enhancer PEE.
What I like about this preprint:
This work approached the process of gastrulation from the viewpoint of cell-intrinsic mechanisms. It showed that when it comes to germ layer specification, epiblast cells are not simply a bystander population obliging to morphogen concentration. Rather, transcriptional regulatory mechanisms allow each cell to be capable of fate decision-making despite the lack of fine-tuned signaling gradients. YAP1, the transcriptional factor in the focus of this study, is mechano-sensitive and has evolutionarily conserved binding sites to key developmental genes. This opens an avenue to study the contribution of in vivo mechanical signaling in the regulation of mammalian gastrulation using mice, thereby circumventing the ethical regulations associated with hESC research. In addition, human-iPSC germ layer specification was shown to be similar to that of hESCs. The knowledge from this work could therefore be adapted to direct differentiation of hiPSCs for effective regenerative therapies.
Questions to the authors:
- YAP1 represses mesendodermal lineages. Given that the edge of 2D gastrulating hESC colonies were mesendodermal, did you observe the nuclear exclusion/down-regulation of YAP1 in the edge cells through immunostaining?
- Genome topology reorganization by YAP1 is reported to be driven by its phase separation [12]. Having said that, did you observe any phase-separated YAP1 foci in the center of the hESC gastruloids?
- YAP1 functions as anti-apoptotic agent in mESCs during differentiation [13]. As cells in YAP1-KO gastruloids appear to be less densely packed, could this be due to increased cell death? Does this reduced density facilitate cell mingling, thereby allowing misplacement of ectoderm lineage?
References
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS and Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–9
- Brennan J, Norris DP and Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–44
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M and Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80
- Arnold SJ and Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G and Pedersen RA. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–49
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R and Thomson JA. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206.
- Moris N, Anlas K, van den Brink SC, Alemany A, Schröder J, Ghimire S, Balayo T, van Oudenaarden A and Martinez Arias A. An in vitro model of early anteroposterior organization during human development. Nature. 2020;582:410–415
- Turner DA, Girgin M, Alonso-Crisostomo L, Trivedi V, Baillie-Johnson P, Glodowski CR, Hayward PC, Collignon J, Gustavsen C, Serup P, Steventon B, P Lutolf M and Arias A. Anteroposterior polarity and elongation in the absence of extra-embryonic tissues and of spatially localised signalling in gastruloids: mammalian embryonic organoids. Development. 2017;144:3894–3906
- Hsu HT, Estarás C, Huang L and Jones K. Specifying the Anterior Primitive Streak by Modulating YAP1 Levels in Human Pluripotent Stem Cells. Stem Cell Reports. 2018;11:1357–1364
- Su Z, Zhang Y, Liao B, et al. Antagonism between the transcription factors NANOG and OTX2 specifies rostral or caudal cell fate during neural patterning transition. J Biol Chem. 2018;293(12):4445-4455. doi:10.1074/jbc.M117.815449
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G and Pedersen RA. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339-49
- Cai, D., Feliciano, D., Dong, P. et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol 21, 1578–1589 (2019).
- LeBlanc L, Lee BK, Yu AC, et al. Yap1 safeguards mouse embryonic stem cells from excessive apoptosis during differentiation. Elife. 2018;7:e40167.
doi: https://doi.org/10.1242/prelights.29810
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Topology changes of the regenerating Hydra define actin nematic defects as mechanical organizers of morphogenesis
Rachel Mckeown
Structural basis of respiratory complexes adaptation to cold temperatures
Pamela Ornelas
Actin polymerization drives lumen formation in a human epiblast model
Megane Rayer, Rivka Shapiro
Also in the cell biology category:
Cell cycle-dependent mRNA localization in P-bodies
Mohammed JALLOH
Control of Inflammatory Response by Tissue Microenvironment
Roberto Amadio
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Bhaval Parmar
Also in the developmental biology category:
Gestational exposure to high heat-humidity conditions impairs mouse embryonic development
Girish Kale, preLights peer support
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Lipid-Based Transfection of Zebrafish Embryos: A Robust Protocol for Nucleic Acid Delivery
Roberto Rodríguez-Morales
preLists in the biophysics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the developmental biology category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)