Pseudo-dynamic analysis of heart tube formation in the mouse reveals strong regional variability and early left-right asymmetry
Posted on: 18 November 2021
Preprint posted on 19 October 2021
Categories: cell biology, developmental biology
Background:
Heart morphogenesis is a complex process that involves a series of changes in cell behaviour and tissue patterning. During gastrulation in mouse embryos, the lateral plate mesoderm splits into two layers, the splanchnic mesoderm located dorsally to the endoderm; and the somatic mesoderm underneath the ectoderm. The primitive heart tube is derived from the splanchnic mesoderm. At the onset of embryonic day 7.5 (E7.5), the cardiogenic mesoderm forms a bilateral field at the anterior region of the primitive streak. The bilateral field merges anteriorly and creates the so-called “cardiac crescent”. The cardiac crescent includes two progenitor pools contributing to the different structures during heart development, including the first heart field (FHF) and the secondary heart field (SHF). From E7.5 to E8.5, cardiac progenitors from two sides migrate toward the midline and coalesce medially to form a single primitive heart tube. From E8.5 onward, the primitive ventricle starts bulging while the anterior and venous poles are elongating. At the same time, the dorsal mesocardium detaches from the dorsal pericardial wall. The primitive ventricle becomes suspended within the pericardial cavity with two poles connecting to the embryo body by the end of these processes. The primitive ventricle gradually loops from left to right, orienting the chamber toward the correct position [1].
Morphometric analysis of tissues is critical for gaining a deeper understand of the molecular mechanism driving organogenesis. However, the morphological changes from cardiac crescent to heart looping occur rapidly and in a brief time window, representing a technical challenge for capturing the spatiotemporal changes of the heart’s structure. Previous methods using serial tissue sections to reconstruct the 3D model produced low-resolution images and were mainly suitable for larger tissue sizes [2]. Here, Esteban et al. study employed a whole-mount imaging technique to scan an entire cardiac region at different developmental stages, establishing new geometrical parameters and generating a high-resolution 3D+t digital model of heart morphogenesis.
Key findings:
1. Image acquisition and segmentation methods:
Mesoderm posterior 1 (Mesp1) is expressed in the nascent mesoderm at the onset of gastrulation and is one of the earliest markers of cardiac progenitors. To visualize tissues derived from the mesoderm lineage, embryos carrying Mesp1-Cre driving the expression of two reporter alleles, R26mTmG and R26Tomato, were used for whole-mount imaging. In these embryos, the mesoderm derived cells were labelled with the green membrane (mGFP) and red cytoplasm (tdTomato); and the remaining cells were distinguished by the red membrane (mTomato). Fifty-two embryos from E7.75 to E8.25 were used to build the tissue atlas, approximately equivalent to one embryo every 20 minutes. The embryos samples were cleared, embedded in 1% agarose gel, and scanned for a whole cardiac region. Depending on the tissue structure, the segmentation proceeded in either automatic or semi-automatic manners. For example, the foregut endoderm was fully automatically segmented, while the myocardium required additional morphological and topological criteria to differentiate from the rest of the splanchnic mesoderm. The segmentation was firstly processed in 2D images, later combined, and then rendered in 3D. The images were subjected to post – segmentation and idealized surface processing to remove noisy voxels and smoothen objects’ surfaces. This methodology successfully reconstructed 3D+t models of the myocardium from cardiac crescent until heart looping, and all mesoderm related tissues including splanchnic, somatic, and paraxial mesoderm and other origin derived tissues such as foregut endoderm and circulatory system.
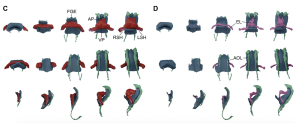
Figure 1: The morphology of the myocardium together with other tissue across stages. FGE, (Foregut endoderm); AP, arterial poles; VP, venous poles; RSH, right sinus horn; LSH, left sinus horn; EL, endocardium lumen; AOL, aortic lumen. Images were obtained from Figure 2 (C-D) in the Esteban I. et al. preprint. DOI: https://doi.org/10.1101/2021.10.07.463475
2. A new morphometric staging system for early heart development:
From E7.75 to E8.25, mouse embryos undergo drastically morphological changes in the structure of the heart tube, foregut socket, head folds and embryo shape. Conventional embryo staging methods relying on the day post coitum (dpc), numbers of somites or head fold shape are therefore not accurate and introduce potential variability in the analysis of heart development. Thus, with 3D reconstructed images, the authors aimed to establish novel morphometric parameters to describe the temporal evolution change of the heart shape more precisely, providing an alternative method for staging embryos.
Based on the anatomical landmarks of the myocardium, mesodermal layers, and foregut, six different measurements were calculated. The first one is the d1/d2 ratio, in which d1 is the length of the border between the myocardium and juxta-cardiac field (JCF) – an anterior tissue above the cardiac crescent, and d2 is the length of the border between the myocardium and secondary heart field (SHF). The second parameter is the proportion between myocardium height to myocardium width (h:w). The third parameter is the ratio between d1 and JCF height (d1:jh). The fourth and fifth are the division of d1 to the foregut length (d1:fl) and the foregut width (d1:fw), respectively. Lastly, the sixth is the ratio between d2 and the height of the somatic mesoderm (d2:sh). To determine the most suitable parameter for staging embryos and the number of embryo groups (k-mean classifier), a k-mean clustering algorithm was applied to each parameter of every embryo. The accuracy of each clustering method was evaluated using average silhouette coefficient (s). Among six parameters, d1/d2 ratio has the best s value and is the most consistent parameter for grouping embryos. Additionally, d1/d2 ratio is well correlated with the heigh-to-width ratio of the heart tube. Based on d1/d2 ratio, embryos could be staged into ten groups in which group 1 – 4 can be assigned to cardiac crescent, group 5 – 8 to heart tube, and group 9 – 10 to heart looping.
3. Standardizing the embryonic geometries of the heart and mapping local shape variability
Given that the heart shapes are highly variable even within the assigned groups, the next aim was to standardize a consensus geometry of the heart tube and the associated tissues in each group and define the local shape variability. The team developed a new quantitative methodology to compute a surface map of each heart sample using equivalent landmark points and curves across stages. Each surface map contains a vertices index for each sample. Then, a vertex-to-vertex correspondence between samples was used to align all heart shapes to a reference shape of the group. All measurements were averaged to create a Mean Shape for each group (MSG). Besides, all shapes in one group were uniformly scaled. Each group selected a medoid shape with the least minimal dissimilarity to be Stage Group Representative (SGR). The concrete set of MSG and SGR is a valuable atlas of the heart structure during heart tube formation and looping. By using MSG to identify the hotspot of natural variability of the cardiac crescent and heart tube, the authors showed that the first morphological variability was found in the bilateral bulges of the cardiac crescent and later at the medial region of the primitive ventricle. At stage 9, when the looping begins, the high morphological variability was also seen in the areas of the out and inflow tracts.
4. Detection of the earliest left-right asymmetry, and 3D + t model of the heart tube formation
Although the left-right asymmetry of the heart is visible at stages 9 and 10 when the heart tube starts to turn rightward, there could be less obvious signs of heart asymmetry happening before the heart looping. Indeed, analysis of 3D quantitative maps of MSG suggested that the angle of insertion of the left and right inflow is not identical. Two measurements were implemented to assess the sinus venous asymmetric structure. One is the φ angle between the direction of inflow insertion and the z-axis, and the other is the θ angle between the orthogonal projection of the direction of the inflow insertion on the XY plane and the x-axis. Then, the changes of these angles were examined relative to the d1/d2 ratio across stages. Interestingly, there was a significant deviation of the θ angle from symmetry initiating at the stages between groups 5 and 6; in contrast, the φ angle on average was unchanged at any stage. Of note, there is an acute angle in the insertion of the right inflow, which is opposite to the smooth transition in the left influx. These results suggested that the left-right asymmetry occurs at a certain angle of the inflow regions.
Finally, the authors used the collection of MSG and SGR to build a temporal dynamic transformation of the myocardium and the associated tissue across ten stages. Tissue meshes from the first shape to the final shape were connected and aligned using vertex coordination. Thirty interpolated shapes were additionally inserted between each stage to smoothen the transition. This approach produces a 3D + t model of heart tube formation, presenting comprehensively morphological changes from heart crescent to heart tube formation and looping.
What I liked about this preprint:
With new embryo imaging techniques and quantitative methodologies, this paper has built an intricate and high-resolution atlas of heart morphology from cardiac crescent to heart tube formation and looping, providing a new set of geometrical parameters for myocardium and the associated tissues during heart development. This atlas not only allows the embryologist to stage embryonic hearts more precisely and minimize the natural variability within embryo samples but also contributes significantly to the studying of congenital heart disease in the aspect of providing morphometric criteria to assess the atypical structure of the different genetically mutated embryonic heart. Finally, the 3D reconstruction method, quantitative analysis, and statistical framework developed in this paper could be applicable to examine the morphogenesis of other organs in mouse embryos or the heart tissue in other species.
Question to the authors
In this preprint, the surface macro-morphology of the myocardium was carefully addressed and reconstructed into a 3D model. It has been known that the myocardial wall of the early embryonic heart tube is also complex that composed of multi-layers with the outer continuous layer and the luminal laminated layer. I wonder if the authors have a plan to build a similar 3D atlas with an exclusive focus on the cross-sectional morphology of the myocardium and whether the same methodology could be applicable to study its structure?
Acknowledgement
I would like to thank Dr Osvaldo Conteras (Victor Chang Cardiac Institute, Australia) and Dr Helen Robertson (preLights Community Manager) for proofreading the highlight.
References
- Kelly RG et al., Heart fields and cardiac morphogenesis. Cold Spring Harb Perspect Med. 2014
- de Boer BA et al., Growth of the developing mouse heart: an interactive qualitative and quantitative 3D atlas. Dev Biol. 2012.
doi: https://doi.org/10.1242/prelights.31061
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Also in the developmental biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Human pluripotent stem cell-derived macrophages modify development of human kidney organoids
Theodora Stougiannou
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the developmental biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)