Isolation of an archaeon at the prokaryote-eukaryote interface
Posted on: 15 August 2019
Preprint posted on 8 August 2019
Article now published in Nature at http://dx.doi.org/10.1038/s41586-019-1916-6
12 years to bring Loki from Asgard to the bench: the first glimpses of the closest living archaeal relatives of eukaryotes
Selected by Dey LabCategories: cell biology, evolutionary biology
Context
Eukaryotes are genetic hybrids. Their genomes appear to have arisen from a merger of archaeal and bacterial genomes some 2 billion years ago. Yet, since both archaea and bacteria tend to have little internal structure, the question remains: how did eukaryotic cell organisation – with a nucleus, organelles linked by a complex transport network, and a highly dynamic cytoskeleton – emerge from such simple prokaryotic origins? This has long remained a mystery, leading to a veritable cottage industry in which researchers try to invent hypotheses about the possible origins of the eukaryotic cell1,2.
The recent discovery, through careful sampling of deep-sea sediments and metagenomic analysis, of a new clade of archaea named for the Asgardian gods of Norse legend promised to shed light on this mystery3,4. One of the most striking discoveries was that the near complete genomes of the Asgard archaea – named Loki, Thor, Heimdall and Odin by Thijs Ettema and colleagues – encode a large number of homologs of Eukaryotic Signature Proteins (ESPs), which include eukaryotic proteins never before detected in prokaryotes: bona-fide actins, actin-binding proteins, tubulin, small GTPases, and a set of domains that are found in proteins with well-studied membrane trafficking functions in eukaryotes. These ESPs helped identify Asgard archaea as the closest living relatives of eukaryotes, and combined with various phylogenetic analyses, have led to a revised view of eukaryogenesis. It now seems clear that the first eukaryotes emerged from within the Asgard clade through a symbiotic association with alpha-proteobacteria5,6. Given the difficulties of inferring phenotype from genotype7, the question remained: what do Asgard archaea actually look like?
Up until last week, no one had definitively cultured, or even seen, an Asgard archaeon. In the absence of cell biology data, one could only speculate, based upon genomic data, about different aspects of Asgard biology such as their metabolism8 or their capacity for phagocytosis9. As a result, most questions about the nature of Asgard archaeal cells remained unanswered. Are Asgard “proto-eukaryotes” with a complex cellular architecture? Do they have a nucleus, internal membrane compartments, and membrane trafficking – or do they look like most other archaea?7 How do they grow and divide?
Key findings
Enter the heroic 12-year culturing effort by Imachi, Nobu and colleagues detailed in this preprint, which provides the first answers to some of the field’s burning questions. To do this, the authors kept marine methane-seep sediments for over 2000 days in an anaerobic methane-fed bioreactor. One of the cultures drawn from the bioreactor became turbid after an additional year of growth. Careful sub-culturing resulted in a stable three-way culture between a Lokiarchaeal strain MK-D1 (the authors suggest the name Candidatus Prometheoarchaeum syntrophicum), Halodesulfovibrio and Methanogenium. Further tweaking – over several years – eliminated the dominant Halodesulfovibrio population and established a final pure co-culture between MK-D1 and Methanogenium. This enabled the authors to sequence the entire Loki genome; the first time this had been done for any Asgard archaeon. This, the authors state (precise details on the overlap are not provided in the preprint), revealed a close match between the published Lokiarchaeum reference and the MK-D1 genome – validating the metagenomic assemblies that led to the original discovery of the Asgard clade.
The paper on MK-D1 includes data on nutrient utilisation and exchange, membrane lipid composition, and cellular ultrastructure (via electron microscopy), which they combined with a genomic and transcriptomic analysis. Rather than restating the conclusions of this technical tour de force in detail, below I discuss a few potential implications of this landmark study. It should be emphasized that many of these have the obvious caveat that other Asgard of the phylum might look very different.
Possible implications for eukaryogenesis: metabolism and lipid chemistry
Genome content, supported by nutrient utilisation data, show that the anaerobic MK-D1 can catabolise amino acids and peptides to produce formate or hydrogen exchanged with a partner species. Given that the other Asgard genomes encode the potential for additional metabolic pathways, notably aerobic respiration in Heimdallarchaeota4, it is worth identifying potentially ancestral features of Asgard metabolism. The authors propose that this ancestor degraded amino acids to produce hydrogen as a byproduct and depended on metabolic partners to complement its limited biosynthetic capabilities, a reconstruction which shares some features with the recent “reverse-flow” model proposed by Spang et al.8 In combination with the fact that MK-D1 appears to only have typical archaeal ether-type membrane lipids, this model has key consequences for models of eukaryogenesis:
- A Loki-like pre-LECA archaeon, an obligate anaerobic symbiont, may have relied on a facultatively aerobic, O2-scavenging partner, the pre-mitochondrial alpha-proteobacterium, to survive aerobic conditions.
- Converting the bacterial partner to a stably propagated endosymbiont would require reconciling two key areas of metabolic overlap – the production of lipids and the generation of ATP from 2-oxoacids.
- If 1 and 2 are correct, they strongly suggest that the process of eukaryogenesis was a slow, ever-closer association evolving over generations7, ruling out the acquisition of the endosymbiont by an acute process such as phagocytic engulfment. This is consistent with the small size of the MK-D1 cell (~550 nm across, from an initial sample of 15 cells) and genomic analyses that deem Asgard archaea incapable of phagocytosis9.
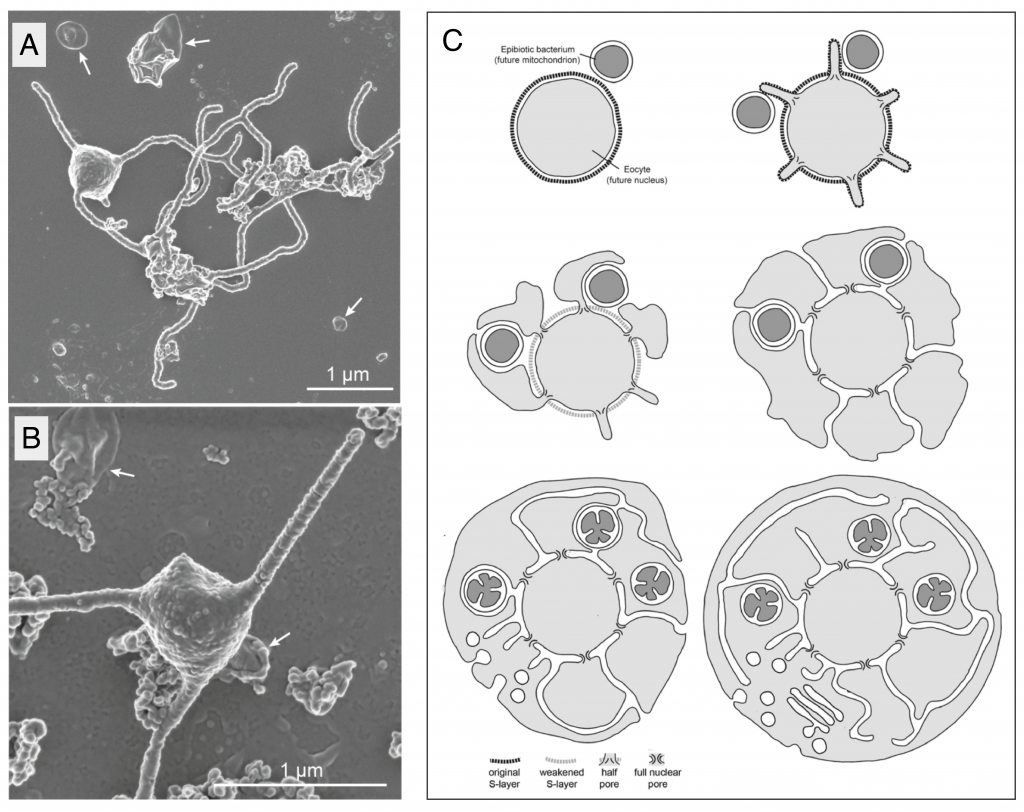
Possible implications for eukaryogenesis: cellular organisation
If the symbiosis was not first established by predatory phagocytosis, how did the pre-mitochondrial partner actually enter the cell, and how and when did the nucleus and endomembrane system evolve? These are the questions that cell biological models of eukaryogenesis must address1.
Here, while remembering that other members of the phylum might look different, MK-D1’s striking shape, with long, branched protrusions emerging from a central bulbous region that appears to contain the DNA (Figure 1 A,B), and complete absence of intracellular membranes could provide a major clue. The authors suggest that a) these protrusions would be likely contact sites for a stable interaction between Asgard archaea and their symbiotic partners, and b) the transition from an ectosymbiotic association to an endocytic one could be achieved by the simple fusion of protrusions that surround the pre-mitochondrial partner. In this “inside-out” topology (Figure 1C), the nuclear envelope is simply the original archaeal plasma membrane and the ER lumen emerges inevitably from the spaces between fused protrusions10.
A model for eukaryogenesis that incorporates these elements was proposed by Baum and Baum in 201410. Excitingly, the central tenet of the “inside-out” model is the capacity of the archaeal host to produce extensive, stable protrusions, like those seen in electron micrographs of MK-D1. The model makes a set of predictions that could be experimentally tested in the future, including the strong prediction that protein complexes serving to stabilise the base of each protrusion in the original archaeal host would give rise to the eukaryotic nuclear pore complex.
Possible implications for the cell biology of Lokiarchaeota
Notwithstanding the impact on models of eukaryogenesis, these first glimpses of a Lokiarchaeal cell also have important consequences for our understanding of the cell biology of this archaeal clade, an exciting new field in its own right. I will discuss a couple of these below.
Small GTPases
The Asgard genomes, MK-D1 included, encode many homologs of eukaryotic small GTPases. In eukaryotes these proteins act as dynamic regulators of membrane trafficking between, and confer identity upon, various internal organelles. However, MK-D1 does not appear to have any internal membranes. Given this observation and the apparent absence from the Asgard genomes of the lipid modification enzymes or consensus sequences required to attach small GTPases to membranes, the small GTPases in this clade must have other functions. These could include functions in nutrient sensing (like Rag GTPases), transport across membranes (Ran GTPase), or in regulating cell shape, morphology and the cytoskeleton – as was previously suggested7 based upon an analysis of the first published Loki genomes.
Actin and actin-modifiers
The MK-D1 protrusions are long, complex branching structures, which suggests a degree of stability. Yet, the EM image of a putative dividing cell (Figure 3c) – suggests they might be lost at division. Therefore, an exciting area of research would be to study how these protrusions are generated, maintained, branched and disassembled. To explore this, a first step would be to generate antibodies against actin and actin-modifying proteins that can be used for immunofluorescence. It will also be important to know the function and dynamics of these protrusions. Are they involved in contacting potential symbiotic partners, such as Halodesulfovibrio or Methanogenium? How do they grow? How are protrusions partitioned at division?
Cell cycle, cell division and ESCRT proteins
MK-D1 expresses archaeal Cdc6 (DSAG12_00518/02854), Mcm (DSAG12_01955) and SMC-complex chromosome partitioning protein (e.g. DSAG12_00806) homologs, much like members of the closely related TACK supergroup of archaea11,12. At first pass, these suggest that MK-D1 might share certain features of the cell division cycle with both TACK archaea and eukaryotes – a replicative phase separated in time from mitosis, and eukaryote-like replicative origin firing. MK-D1 also expresses the prokaryotic tubulin homolog FtsZ (DSAG12_00639/01941) as well as components of the ESCRTII and ESCRTIII protein complexes. Does FtsZ drive the division of these cells at the end of the cell cycle (in a similar way to many other archaeal and bacterial clades), or is cell division performed by the ESCRT complex – as it does in TACK family archaea13–15? Or are both FtsZ and ESCRTIII involved – as appears to be the case in many eukaryotes? If ESCRT is not involved in cell division in these Asgard archaea, perhaps the ESCRT pathway is responsible for the trafficking of ubiquitinated substrates into vesicles (e.g. extracellular vesicles – as proposed for TACK archaea)16? Given the relatively large genome and small cellular volume, how is the genome organised and/or compacted, and is this cell-cycle-dependent? Does MK-D1 have histones?
Other interesting potential ESPs in the MK-D1 transcriptome
In addition to the ESPs detailed in the Asgard discovery papers, there are a few annotations in Supplementary Table S6 that would be of broad interest if validated:
Leucine-rich repeat-containing G-protein coupled receptor 6 (DSAG12_02079; non-canonical GPCR, part of a signalling pathway thought to be eukaryote-specific17, in humans Q9HBX8)
Laminin (DSAG12_02394; involved in ECM-binding in metazoans, in humans Q16787)
Programmed cell death protein 5 (DSAG12_03236; apoptosis, in humans O14737– cell death machinery is currently thought to have been inherited from bacteria18)
COP9 signalosome subunit 5 (DSAG12_03674; regulator of ubiquitin conjugation, in humans Q92905)
Transitional endoplasmic reticulum ATPase (DSAG12_01511; remodeling of ER and Golgi during mitosis, in humans P55072)
Scribble (DSAG12_02613; metazoan scaffold protein involved in apico-basal polarity, in humans Q14160)
Conclusions
There is no doubt that this landmark study will have a lasting impact on the fields of archaeal cell biology and eukaryogenesis. The authors are to be commended for the incredible amount of careful, patient work that led to this breakthrough, as well as for openly sharing these findings with the research community immediately by preprinting. We also hope the community will have a chance to analyse the full genomic sequence in detail soon. Thrilling times lie ahead, as more Asgard archaea are gradually brought out of the shadow lands into the lab – and into realm of experimental exploration.
Further reading
Perhaps unsurprisingly, this preprint has already generated waves on social media and traditional media outlets! Take a look at these news articles in Science, Nature and New Scientist, or start with this Twitter thread from Thijs Ettema.
Acknowledgements
I am grateful to David Baum, Buzz Baum, Del Mutavchiev, André Pulschen and Diorge Souza for discussions and critical comments on this highlight, to the authors for comments on technical accuracy, and members of the preLights team for proof-reading.
References
- Baum, D. A. A comparison of autogenous theories for the origin of eukaryotic cells. Am. J. Bot. 102, 1954–65 (2015).
- Martin, W. F., Garg, S. & Zimorski, V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140330 (2015).
- Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
- Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
- Wang, Z. et al. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 5, 7949 (2015).
- Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018).
- Dey, G., Thattai, M. & Baum, B. On the Archaeal Origins of Eukaryotes and the Challenges of Inferring Phenotype from Genotype. Trends Cell Biol. 26, 476 (2016).
- Spang, A. et al. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat. Microbiol. 4, 1138–1148 (2019).
- Burns, J. A., Pittis, A. A. & Kim, E. Gene-based predictive models of trophic modes suggest Asgard archaea are not phagocytotic. Nat. Ecol. Evol. 2, 697–704 (2018).
- Baum, D. A. & Baum, B. An inside-out origin for the eukaryotic cell. BMC Biol. 12, 76 (2014).
- Lundgren, M. & Bernander, R. Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104, 2939–44 (2007).
- Lindås, A.-C. & Bernander, R. The cell cycle of archaea. Nat. Rev. Microbiol. 11, 627–38 (2013).
- Makarova, K. S., Yutin, N., Bell, S. D. & Koonin, E. V. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8, 731–41 (2010).
- Lindås, A.-C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J. G. & Bernander, R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U. S. A. 105, 18942–6 (2008).
- Pelve, E. A. et al. Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol. Microbiol. 82, 555–566 (2011).
- Ellen, A. F. et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13, 67–79 (2009).
- de Mendoza, A., Sebé-Pedrós, A. & Ruiz-Trillo, I. The Evolution of the GPCR Signaling System in Eukaryotes: Modularity, Conservation, and the Transition to Metazoan Multicellularity. Genome Biol. Evol. 6, 606–619 (2014).
- Koonin, E. V & Aravind, L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 9, 394–404 (2002).
doi: https://doi.org/10.1242/prelights.13336
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Tissue mechanics and systemic signaling safeguard epithelial tissue against spindle misorientation
Ruoheng Li
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Also in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (1 votes)
(1 votes)