CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis
Posted on: 21 July 2023 , updated on: 4 August 2023
Preprint posted on 26 June 2023
Proteasomal misprocessing for early onset of tauopathies: A CRISPRi screen identifies how cellular environment dictates tau proteostasis
Selected by Kritika MehtaCategories: cell biology, neuroscience
Background:
Protein aggregation is one of the leading causes of various neurodegenerative disorders1. In this preprint, the authors focus on a widely studied intrinsically disordered protein called Tau. Tau aggregation gives rise to a family of disorders collectively known as tauopathies. Since most of the tauopathies are acquired in adulthood, factors in the cell milieu govern disease onset and progression. However, early onset drivers of tau aggregation and their mechanistic involvement in disease pathogenesis have not been studied extensively. For example, GWAS studies have uncovered various risk factors but do not provide any information on molecular mechanisms. Additionally, sc-RNA seq can identify differential expression of genes between diseased and normal state but cannot pinpoint the exact role in pathology or early onset.
The focus of this manuscript was to identify the key factors underlying the selective vulnerability of tauopathies among different neuronal cell types. The iPSC-derived neurons used in this study provide an amenable model system for this study since they are physiologically relevant and allow for high-throughput screening experiments. Understanding the specific role of the cellular environment will also help to explain the selective vulnerability of different neuronal cell types to tau aggregation.
Key findings:
Model optimization for high throughput genomic screening
To establish a model system for detecting tau oligomers, the authors examined WT and V377M mutant of tau in iPSC-derived neurons. Neurons were stained for tau oligomers using a T22 antibody, and cells were screened for level of oligomerization using flow cytometry. Neurons expressing both a single and double copy of V377M mutant, showed a significant increase in the accumulation of aggregates compared to WT (see fig.1).
The CRISPRi screens
The authors then knocked down different genes in V322M expressing neurons and sequenced the neurons sorted for different levels of tau oligomerization. This knockdown screen identified various components that were already known to decrease tau oligomer levels (examples: autophagy modulators2 and m6A regulatory genes3). One important hit from the screen – showing increased tau oligomer levels – involved the Electron transport chain (ETC). Interestingly, Ubiquitin proteasomal system (UPS)-related factors were also among the top hits, as well as those relating to the GPI-anchor biosynthesis. Additionally, Melvanote – a precursor of cholesterol – decreased tau levels and genes essential for UFMylation, already known to control pathogenic tau levels, also showed up as a hit.
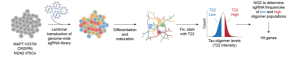
Figure 1 Original(2A) Schematic of the CRISPRi screen performed on iPSC-derived neurons to test the effect on tau oligomerization.
To characterize the top hits more closely, 1,037 genes were pooled into a secondary sgRNA library. The authors then screened both WT and V377M mutant using a total tau antibody and just the WT using T22 antibody. From this, they could conclude that the WT tau screen seemed to be more sensitive to the knockdown of genes involved in mRNA transport, while the V377M screen was enriched for hits regulating the mTOR signaling pathway. Cul5 – an E3 ubiquitin ligase – showed up for both WT and mutant screens. Since Cul5 is highly expressed in excitatory neurons which are relatively resilient to Alzheimer’s disease (AD) 4, the authors pointed out that this might contribute to the selective vulnerability of cell types in AD progression.
What is the role of the proteasomal machinery in tau oligomerization?
Since Cul5 and its regulator RNF7 were strong hits in the screen, the authors over-expressed Cul5 and tau in cells to test if they physically interact. Immunoprecipitation revealed that tau co-precipitated with Cul5. RNF7 was also present confirming it to be a functional protein assembly. Now to identify the region in tau recognized by Cul5, supposedly a degron, a number of sub-regions of tau were expressed in the neurons. The authors were able to identify a region (amino acids 80-130) that is recognized by Cul5. They also identified that Cul5 interacted with tau using the adaptor protein SOCS4.
What is the effect of mitochondrial dysfunction on tau oligomerization?
To further their understanding of how the cellular environment promotes tau interaction with the proteasomal machinery, the authors moved their attention to the mitochondrial genes that showed up in the screen. These included FECH, PSAP and FH and interestingly suppression of all these genes increased ROS level in the neurons. The authors then checked how mitochondrial function might be controlling tau oligomer levels by using a pharmacological approach. They could conclude that ROS production is a side-effect of mitochondrial dysfunction which induces tau misprocessing.
To support their findings, the authors induced oxidative stress in the neurons with hydrogen peroxide treatment. Oxidative stress increased tau misprocessing into a 25kDa fragment. Increasing expression of the proteasomal activator PA.28 reduced the levels of the 25kDa tau fragment as expected, suggesting it is the inability of the proteasome to successfully degrade the oxidized proteins during oxidative stress that leads to the accumulation of misprocessed tau fragments.
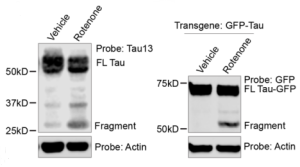
Figure 2 Original (5C, 5D) Effect of Rotenone on endogenous Tau fragmentation (left). Effect of Rotenone on exogenous GFP-Tau fragmentation in iPSC-derived neurons
Is tau oxidation the cause of its misprocessing?
The authors hypothesized that the N-terminal methiones of tau were oxidized during stress. To test if oxidized tau levels lead to misprocessing of tau every methionine was mutated to leucine. They observed that, with mutant tau expression levels being the same as WT, there was a large decrease in mutant tau fragmentation. Thus, direct tau oxidation during stress forces tau into the proteasome for aberrant degradation.
What is the sequence of the misprocessed tau fragment?
Lastly, to identify the tau sequence created after being misprocessed by the proteasome, the authors purified tau from neurons and digested it with GluC to mimic the proteasome. The sequences were then identified using LC-MS/MS and narrowed down to 9 residues (172-200). This was consistent with the tau biomarker in CSF patients that ends near residue 2305. The authors could conclude that N-terminal tau fragments may serve as markers for neuronal oxidative stress that results in changes in proteasomal processivity.
What I liked about the preprint:
I am interested in neuroscience and specifically in understanding how the intracellular environment becomes supportive of disease progression. Understanding the origin and early markers of neurodegeneration can help understand the overall development of diseases into later stages. Proteins like tau have been studied extensively for their involvement in the progression of neurodegenerative diseases. This manuscript highlights how neurons become vulnerable when the cellular machinery is compromised due to internal and external factors leading to oxidative stress. Mitochondrial dysfunction is an important factor that allows the opportunistic misprocessing of tau protein which leads to uncontrolled aggregation in later disease stages. The authors’ CRISPR-based strategy in iPSC-derived neurons allows for high throughput screening in a physiologically relevant system.
Questions for the authors:
- As discovered here, oxidative stress can lead to tau fragmentation due to misprocessing. Is it known which cell types in particular are more likely to be affected as they could serve as the sites of onset of tauopathies?
- Regarding the proteasome recognition (degron-like) sequence of tau identified in this manuscript, is it known to be mutated in any tauopathies currently known?
- Are residues of the proteasomal machinery-or even-Cul5 also oxidized (which could be the reason for its aberrant processing of tau)?
Future Directions:
Future studies should focus on the effects of early onset tau misprocessing on the propagation of action potentials in the axons. This would provide further insight into cell type vulnerability to protein aggregation eventually causing neurodegeneration. It will also be interesting to see what other protein fragments accumulate due to the misprocessing of the proteasome.
References:
- Ross, C. A. & Poirier, M. A. Protein aggregation and neurodegenerative disease. Nat Med 10, S10–S17 (2004).
- Caballero, B. et al. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat Commun 12, 2238 (2021).
- Jiang, L. et al. Interaction of tau with HNRNPA2B1 and N6-methyladenosine RNA mediates the progression of tauopathy. Mol Cell 81, 4209-4227.e12 (2021).
- Leng, K. et al. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat Neurosci 24, 276–287 (2021).
- Cicognola, C. et al. Novel tau fragments in cerebrospinal fluid: relation to tangle pathology and cognitive decline in Alzheimer’s disease. Acta Neuropathol 137, 279–296 (2019).
doi: https://doi.org/10.1242/prelights.35154
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Also in the neuroscience category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)