3D genome organization around nuclear speckles drives mRNA splicing efficiency
Posted on: 16 February 2023 , updated on: 29 May 2024
Preprint posted on 4 January 2023
Article now published in Nature at http://dx.doi.org/10.1038/s41586-024-07429-6
mRNA in the right place, at the right time: a new preprint explores how dynamic organization of genomic DNA around nuclear speckles determines mRNA splicing efficiency.
Selected by Diego MartínezCategories: cell biology, molecular biology
Updated 29 May 2024 with a postLight by Diego Martínez
The work by Prashant Bhat and collaborators led by Mitchell Guttman has now been published in Nature.
I have noticed a great similarity between the published and preprinted version of this work in terms of the main figures and text, which speaks of the quality of this work. The extended figures suggest that the focus of the review process was to strengthen the authors’ conclusions by adding experimental controls or variables. For example, to dissect the efficiency of splicing in different experiments additional variables were taken into consideration, such as gene expression levels or genomic features (e.g., gene length), even though the authors managed to successfully recapitulate their results using reporters. Another example would be related to the use of these reporters, as the authors now show the reproducibility of location relative to speckles of integrated reporters in comparison with endogenous loci.
Since the main message of the paper has remained unchanged, one can say that the model presented in this work is still standing strong and that the experiments requested as part of the peer review process have only strengthened it.
Congratulations to the authors.
Background
The nucleus is a highly ordered cellular compartment where proteins and nucleic acids display a distinct spatial organization. In mammalian cells, several nuclear bodies can be distinguished, including the nucleolus, Cajal Bodies or nuclear speckles, which constitute compartments of a particular composition.
Nuclear speckles are domains enriched in pre-mRNA splicing factors. Although they have been suggested to be sites of active mRNA splicing, there is a lack of evidence for the direct implication of these structures in the catalytic process. A long-standing view of nuclear speckles is that they merely serve as storage hubs for splicing factors, a notion suggested by observations such as the diffusion of these proteins from speckles as a result of post-translational modifications(1-4).
On the other hand, a hypothetical functional role of these nuclear bodies in mRNA processing stems from indirect evidence such as alterations in nuclear speckle morphology upon transcription or splicing inhibition(5,6) or conversely, the defects in splicing arising from interfering with nuclear speckle formation(7). Moreover, the accumulation of poly-A rich transcripts and spliced mRNAs at the speckle periphery strongly suggests that co-transcriptional splicing occurs in the nuclear speckles’ vicinity (discussed in 8).
Previous work from this and other groups has shown that specific genomic regions contact nuclear bodies, including nuclear speckles(9,10,12). By demonstrating the functional implications of these contacts, this preprint sheds light on the influence of nuclear speckles in genome organization and mRNA splicing efficiency.
Key findings
Highlights:
- Speckle-close pre-mRNAs have a higher concentration of spliceosome components.
- Efficiency of splicing correlates with gene locus proximity to nuclear speckles independently of genomic features.
- RNA Pol II density determines the interaction of a genomic region and nuclear speckles, imposing cell type-specificity based on transcriptional programs.
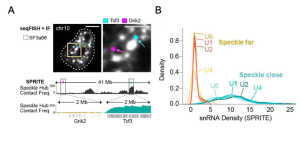
The authors leveraged their own method to explore 3D genome organization and RNA processing: SPRITE(10). Briefly, with this technology proteins and nucleic acids are crosslinked, nucleic acids are barcoded and then sequenced. Together with a complementary immunofluorescence-based approach (seqFISH+), the authors show that nuclear speckles contact specific genomic regions (Figure 1A).
Suggesting that these contacts could have implications beyond genomic DNA organization, they show that regions close to nuclear speckles present higher spliceosome concentrations as measured by small nuclear RNA (snRNA) levels (Figure 1B), in mouse embryonic stem cells (mESCs).
Higher spliceosome binding to speckle-close pre-mRNAs could hypothetically lead to increased co-transcriptional splicing efficiency. To analyze this, the authors focus on splicing of pre-mRNA that occurs near the locus from which it is transcribed (co-transcriptional splicing), by using methods that allow measurement of mRNAs associated with chromatin. Remarkably, they report differences in splicing ratios depending on pre-mRNA proximity to nuclear speckles as high as 3-fold.
Figure 1. Genomic regions in contact with nuclear speckles are enriched in snRNAs. A) Reconstructed image for DNA seqFISH+ and immunofluorescence (SF3A66-nuclear speckle marker) in mESCs comparing a speckle close gene (Tcf3, blue) and speckle far gene (Grik2, purple) (top). Speckle contact frequencies from SPRITE for chromosomes 10 at 100-kb resolution (bottom). Zoom in, speckle contact frequencies from SPRITE for the 2 Mb region around genes shown in top. B) Normalized density of U1, U2, U4, U6 snRNAs on speckle close versus speckle far genomic regions. Normalization for each snRNA is to the mode of the speckle far distribution to visualize all snRNA densities on the same scale. RPKM for both speckle far and close genes is thresholded between 2.5-7.5.
To exclude the influence of genomic features on these differences (e.g., gene length), the authors generated a GFP splicing reporter with an intron that prevents GFP expression (Figure 2). In addition, this reporter contained an MS2 bacteriophage RNA hairpin within the intron that binds with high affinity to MS2 coat protein (MCP). By co-expressing the splicing reporter together with specific mCherry-MCP-fusion proteins known to localize at different locations within the nucleus (Figure 2 and Figure 3A), they could study splicing efficiency relative to nuclear speckle proximity. Their results indicate that recruitment of a pre-mRNA to nuclear speckle proteins, but not to other nuclear proteins, is sufficient to increase, in a non-linear manner, splicing efficiency (Figure 3A and 3B).
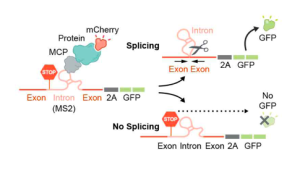
Figure 2. Schematic of pre-mRNA splicing assay via a fluorescence based read out.
Emerging evidence that the catalytic spliceosome is physically close to RNA Pol II during co-transcriptional splicing(11) suggests that the contacts between genomic DNA and nuclear speckles may facilitate this phenomenon. RNA Pol II occupancy analysis revealed that indeed, genomic regions proximal to speckles were enriched in RNA Pol II compared to distant regions.
Remarkably, a comparison of SPRITE maps generated in mouse myocytes to the ones previously created in mESCs, revealed that genome organization around nuclear speckles was cell type specific. Although many genomic regions were nuclear speckle proximal in both cell types, the remaining areas were unique. Consistent with the correlation between RNA Pol II occupancy and nuclear speckle proximity, differential genomic positioning reflected cell type specific characteristics. For example, the pluripotent-specific gene (Nanog) is speckle proximal only in mESCs, whereas a skeletal muscle-specific gene (Ttn) was close to speckles exclusively in myocytes (Figure 3C). A similar result was obtained for the comparison between distinct human cell types.
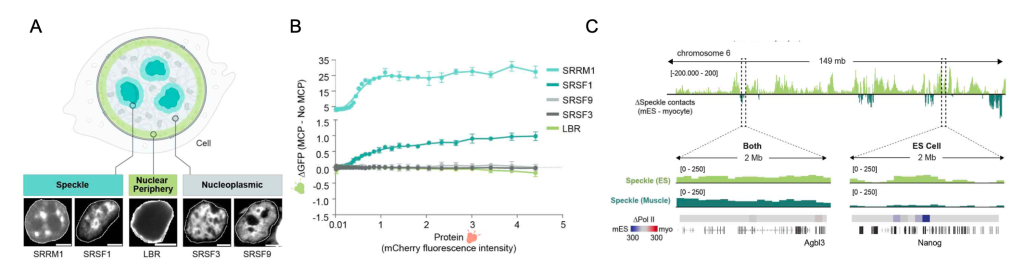
Figure 3. Co-transcriptional splicing efficiency varies based on cell type-specific proximity to nuclear speckles. A) Localization of proteins used in mCherry fusion protein reporters. B) Difference in splicing efficiency based on GFP expression between constructs with MCP and no MCP versus mCherry fluorescence intensity (x axis) for all fusion proteins tested. C) Difference in speckle hub contact frequency between mESCs and myocytes (chromosome 6) and 2-Mb zoom in regions of speckle contact frequencies and Ser2P Pol II densities for Agbl3 (speckle in both) and Nanog (mESC specific).
Why I like this preprint
I became interested in this preprint because it addresses a long-standing question (functional role of nuclear speckles in splicing), from the innovative perspective of genomic DNA organization. The authors propose a model in which high splicing factor concentration at nuclear speckles, with elevated affinity for pre-mRNA, drives RNA pol II (pre-mRNA)-dense regions to the speckle’s periphery, shaping genome organization and acting as splicing catalyzers. In other words, shaping genome organization through transcription (high RNA Pol II density) would rely on nuclear speckles (high splicing factor density). This provides functional significance to the observation of chromosomal-speckle contacts and the concept of ‘’paraspeckle’’. Taken together, these results are fundamental to our understanding of the physiological relevance of nuclear bodies in RNA processing and genome organization.
As the authors of this preprint propose, this feature may represent an additional level of gene expression regulation. I believe this notion may be essential to characterize the role of several splicing factors which display dynamic re-localization (e.g., SR proteins), in time and space. For example, it could reveal important aspects of splicing modulation in specific cell types at different developmental stages. Moreover, exploring nuclear positioning of splicing factors–i.e., nuclear speckle composition–may help to precisely determine their influence in pathogenic variant expression beyond their mutational status and expression levels.
Considering the cell type-specific nature of these contacts, and related to my field (molecular oncology), I believe this feature may potentially shape transcriptome diversity in the context of cancer cell heterogeneity, thereby possibly influencing mechanisms of drug resistance and processes like metastasis or immune escape.
References
- Matta, I. Splicing in space. Nature 372, 727–728 (1994). doi.org/10.1038/372727a0
- Zhang, G., Taneja, K., Singer, R. et al.Localization of pre-mRNA splicing in mammalian nuclei. Nature 372, 809–812 (1994). doi.org/10.1038/372809a0.
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996 Jan 15;15(2):265-75. org/10.1002/j.1460-2075.1996.tb00357.x
- Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999 Oct;19(10):6991-7000. doi: 10.1128/MCB.19.10.6991.
- Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996 Oct;7(10):1559-72. doi: 10.1091/mbc.7.10.1559.
- O’Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol. 1994 Feb;124(3):249-60. doi: 10.1083/jcb.124.3.249.
- Kurogi Y, Matsuo Y, Mihara Y, Yagi H, Shigaki-Miyamoto K, Toyota S, Azuma Y, Igarashi M, Tani T. Identification of a chemical inhibitor for nuclear speckle formation: implications for the function of nuclear speckles in regulation of alternative pre-mRNA splicing. Biochem Biophys Res Commun. 2014 Mar 28;446(1):119-24. doi: 10.1016/j.bbrc.2014.02.060.
- Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006 Jul;288(7):664-75. doi: 10.1002/ar.a.20336.
- Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014 May 19;24(10):1138-44. doi: 10.1016/j.cub.2014.03.053.
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, Cai L, McDonel P, Garber M, Guttman M. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 2018 Jul 26;174(3):744-757.e24. doi: 10.1016/j.cell.2018.05.024.
- Herzel, L., Ottoz, D., Alpert, T. et al.Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol 18, 637–650 (2017). https://doi.org/10.1038/nrm.2017.63.
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999 Feb 22;144(4):617-29. doi: 10.1083/jcb.144.4.617.
doi: https://doi.org/10.1242/prelights.33728
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
Also in the molecular biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
preLists in the cell biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)