Quantification of gallium cryo-FIB milling damage in biological lamella
Posted on: 8 March 2023
Preprint posted on 3 February 2023
In this preprint, the authors quantify the damage caused by cryo-FIB milling to biological specimens and how this information can be leveraged to prepare samples for in situ structure determination.
Selected by Kanika KhannaCategories: biophysics, cell biology
Background
In situ cryo-electron tomography (cryo-ET) is emerging as a promising technique to determine the structure and function of macromolecular complexes inside their native environment, i.e., the cell, to near-atomic resolution. But only thin cells (typically less than ~500 nm) are amenable to cryo-ET and most thicker biological samples require thinning to make them electron-transparent. Cryo-focused ion beam (cryo-FIB) milling is now widely used to thin biological specimens by using a focused beam of gallium ions to ablate cellular material. Thin sections of cells produced by this method, called lamellae, are typically in the range of 100-300 nm which are later processed for cryo-ET data acquisition and subsequent subtomogram analysis.
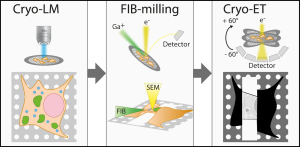
Schematic showing cryo-ET workflow for a mammalian cell imaged by a cryo-light microscope, followed by milling inside a cryo-FIB and subsequent imaging using tilt-series acquisition. Image adapted from Klein et al., 2021 that is made available under a CC-BY 4.0 license.
Much has been reported on the radiation damage caused when an electron beam interacts with the sample during tilt-series acquisition inside a transmission electron microscope (TEM). But we know little about the sample damage caused by gallium ions during FIB-milling which may interfere with high-resolution reconstructions. Previous work has reported that at conditions used for cryo-lamellae preparation, gallium ions are deposited in the sample at a depth of ~10-20 nm from the lamella surface and collisions between gallium ions and sample atoms may lead to additional damage affecting the sample integrity and quality. In this preprint, the authors set out to explore the nature of the damage introduced by gallium ions during cryo-FIB milling and quantify it so that researchers can guide the process of lamellae production for favorable in situ cryo-ET analysis.
Key Findings
Matching template to target to figure out the damage
Previously, the authors had developed an approach called 2D template matching (2DTM) to locate macromolecular complexes in 3D from 2D cryo-electron microscopy images of either unmilled or FIB-milled cells. Individual target complexes from these images can then be compared with a high-resolution template from a molecular model. The similarity is reflected by a parameter called the 2DTM signal-to-noise ratio (SNR). Damage caused by FIB-milling to the sample and subsequently to target molecules would then decrease the correlation between the template and the target, resulting in a lower 2DTM SNR.
In this study, the authors performed cryo-FIB milling to visualize the cytoplasm of yeast Saccharomyces cerevisiae where ribosomes are present in high density. This system provides a platform to test target (large ribosomal subunits or LSU) integrity across the lamellae and assess sample damage caused by FIB-milling using 2DTM.

Template generated from the atomic coordinates of mature 60S ribosomes is mapped onto a cryo-EM micrograph of yeast nuclear periphery of a FIB-milled lamella. 2DTM SNR is represented such that peaks corresponding to significant template/target match can be demarcated from background. Image adapted from Lucas et al., 2022 that is made available under a CC-BY 4.0 license.
How does damage vary across the lamella depth?
By making lamellae of different thicknesses, the authors found that the 2DTM SNRs of LSUs were lower at lamellae edges compared to the center. The authors then quantified the depth of the damage in ~200 nm lamellae as at this thickness, LSUs were distributed uniformly throughout the lamellae and the SNR was constant at the center where there is likely a stretch of minimal FIB-milled damage. The authors found that for distances ≤60 nm from the lamella surface, the relative 2DTM SNR was significantly lower compared to >60 nm, suggesting a loss of target integrity due to FIB-milling damage. They also found the nature of this damage to be that of exponential decay, with the first ~10-20 nm from the lamella surface suffering the most damage. As a control, they also looked at unmilled Mycoplasma pneumoniae cells and did not notice any such damage.
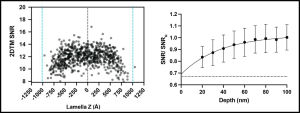
Scatterplot showing the 2DTM SNR of each LSU as a function of z-coordinate relative to the lamella center for a 200 nm lamella (left) and the mean change in SNR at indicated lamella depths relative to the surface. Image adapted from Lucas et al., 2023 that is made available under a CC-BY 4.0 license.
What is the nature of FIB-milling damage?
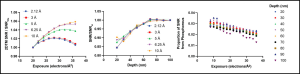
The authors then decided to tackle the mechanism of FIB-milling damage and asked if it is similar to radiation damage caused by electrons during cryo-EM data acquisition. Radiation damage affects signals at high spatial frequencies which in turn contributes to the mismatch between the target and the template structures. Due to this, cryo-EM is performed in low dose conditions to minimize sample exposure to the electron beam. To assess the radiation damage, the authors low-pass filtered the template to different spatial frequencies and calculated the change in 2DTM SNR of LSUs under different electron exposure conditions. They found that for templates filtered to lower spatial frequency (between 1/10 and 1/7 Å-1) the SNR increased with increasing exposure, but at higher spatial frequencies the SNR decreased with increasing exposure, consistent with previous findings. However, the SNR for FIB-milled samples did not vary by lamella depth and remained almost constant at the lamella center for different spatial frequencies above 1/5 Å-1, suggesting some sort of permanent damage to the target structure during milling.
In addition, the authors found that the SNR contributed by phosphorous atoms in structures decreased faster with increasing electron dose compared to the loss from the overall structure, consistent with the idea that radiation damage causes breakage of phosphodiester bonds. However, the decrease in SNR was consistent at different lamella depths, suggesting FIB-milling damage is caused by a mechanism that is different from radiation damage caused by electrons.
Plots showing change in 2DTM SNR as a function of electron exposure and relative lamella depth for templates filtered to different resolutions, and loss of SNR due to damage to phosphorus atoms in target structures at different lamella depths. Image adapted from Lucas et al., 2023 that is made available under a CC-BY 4.0 license.
How much or little to mill, then?
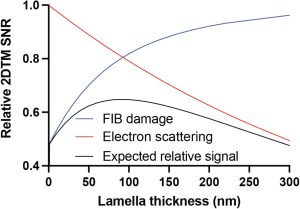
“How much to mill?” is almost always the hard question when cryo-FIB milling. Thicker lamellae have more biological information but suffer from signal loss due to inelastic electron scattering, resulting in noisy images. On the other hand, thinner lamellae have less information but offer the possibility of obtaining high-resolution structures with less background noise. Combining this with the new information presented in this preprint that FIB-milling can introduce damage in ~60 nm from each lamella surface, leaves one with only so much room to make samples with minimal FIB-milling or radiation damage. The authors developed a model to assess the relative impact of FIB-milling damage and lamella thickness on high-resolution imaging. The model predicts that for lamellae thinner than 90 nm, the FIB-milling damage dominates but for lamella thicker than 90 nm, FIB-milling damage is less significant compared to signal loss due to electron scattering.
The expected loss in signal due to either FIB-milling damage as a function of lamella thickness is less than that of electron scattering in thicker samples. Image adapted from Lucas et al., 2023 that is made available under a CC-BY 4.0 license.
What I liked about the preprint
Overall, the preprint presents an important advance in our understanding of potential sample damage caused during cryo-FIB milling of biological specimens. Thinking of potential solutions to negate the damage can result in the detection of more targets as well as preserve their integrity better to determine high-resolution in situ structures. The preprint also highlights the use of 2DTM SNR to evaluate sample integrity during cryo-EM imaging.
The information presented in the preprint is important for researchers working at the interface of both cryo-EM method development and structural cell biology. I appreciated that the authors present their findings in a manner that could be appreciated by both sets of researchers and bridges the gap between the two fields.
Questions for the author
- Do the authors think the FIB current for fine milling influences the depth of damage? There is always a debate about whether to do fine polishing at 10 pA for longer or 30 pA for shorter times. Also, thinner bacterial cells mill faster than thicker mammalian cells. Does the fact that the final polishing steps are shorter in some specimens compared to the others influence the depth of damage?
- What are the authors’ views on FIB voltages lower than 30 keV on the depth of damage? Researchers in the material sciences field have shown that lower FIB voltages result in lower sidewall damages and sometimes combining low voltage FIB with Argon ion beam can also remove potential damage. Is damage removal feasible in cryo-FIB milling at all?
doi: https://doi.org/10.1242/prelights.34034
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
ROCK2 inhibition has a dual role in reducing ECM remodelling and cell growth, while impairing migration and invasion
Sharvari Pitke
Also in the cell biology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Taxane-Induced Conformational Changes in the Microtubule Lattice Activate GEF-H1-Dependent RhoA Signaling
Vibha SINGH
PIP5K-Ras bistability triggers plasma membrane symmetry breaking to define cellular polarity and regulate migration
Vibha SINGH
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)