Tight nanoscale clustering of Fcγ-receptors using DNA origami promotes phagocytosis
Preprint posted on 18 March 2021 https://www.biorxiv.org/content/10.1101/2021.03.18.436011v1.full
Article now published in eLife at http://dx.doi.org/10.7554/elife.68311
Categories: cell biology
Background
Phagocytosis (from Ancient Greek (phagein) ‘to eat’, and (kytos) ‘cell’) is the process by which cells consume foreign particles from their environment and plays an essential role in humoral immunity (Fig. 1A). Macrophages recognize invading pathogens like bacteria and viruses after they have been identified and bound by antibodies such as IgG. This binding of pathogens by IgG is called opsonization. Once macrophages encounter an antibody-opsonized target, receptors on their cell surface bind to the Fc region of the IgG antibody. When macrophages have bound to a significant number of IgG molecules, they undergo extensive morphological changes to envelop and consume the opsonized target (Fig. 1A). A big question in the field is what constitutes a significant number of IgG molecules; what are the rules for phagocytosis?
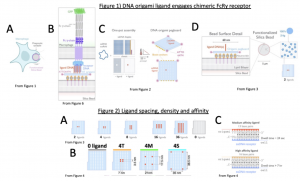
A key macrophage receptor that recognizes the Fc region is the Fc gamma receptor (FcɣR, Fig. 1B). Binding of FcɣR to Fc initiates an intracellular signaling cascade resulting in cytoskeletal and membrane protein rearrangements that facilitate phagocytosis. Previous studies have demonstrated that increased FcɣR engagement promotes phagocytosis, but the optimal spacing and density of engaged FcɣRs remains a major open question in the field. In this preprint, Kern et al. address this question with a synthetic biology approach. They make a chimeric FcɣR molecule that can bind to DNA oligonucleotide ligands and combine this approach with a new DNA origami-based method that allows them to precisely control the spacing and arrangement of these oligonucleotide ligands (Fig. 1B). This method allows them to exactly determine the optimal spacing and density of FcɣR ligands required to trigger phagocytosis.
DNA origami uses single stranded DNA and “staple oligos” to anneal a DNA template into a three-dimensional structure of precise shape and orientation. In this study the authors are able to create a rectangular DNA origami “pegboard” onto which the oligonucleotide ligands can be arranged with nanometer precision and anchored to silica beads (Fig. 1C, 1D). The authors use these ligand-opsonized beads to engage FcɣR receptors in human macrophages in various arrangements and densities, identifying optimal receptor positioning for phagocytosis of these synthetic particles.
- The authors engineered macrophages with synthetic Fcɣ receptors that could be attached to DNA oligonucleotides (DNA-CARɣ). Binding to DNA-ligands can induce clustering of these DNA-CARɣ molecules (Fig. 1A).
- They combined these cells with silica beads coated with complimentary DNA oligonucleotide ligands that were precisely arranged using DNA-origami technology (Fig. 1C, 1D). Annealing of the oligo-ligands to the DNA-CARɣ molecules mimics the binding of an Fc region to the endogenous FcɣR and initiates phagocytosis.
- The DNA Origami platform is called a “pegboard” (analogous to a pegboard where tools can be hung in many different ways (Fig. 1C, 1D). Oligo-ligands arranged on the pegboard platform were used to activate DNA-CARɣ macrophages and allows manipulation of ligand spacing at the nanometer-resolution to quantitatively investigate receptor engagement and downstream signaling pathways (Fig. 2A, B).
Summary Figures (adapted from Kern et al)
Key Findings
- A cluster size of 8 ligands is the critical density threshold for FcɣR clusters needed to initiate FcɣR signalling and phagocytosis.
- The strongest initiator of phagocytosis was the closest possible clustering of receptor-ligands (3.5 nm apart, Fig. 2A) on the particle. This experiment also suggested that avidity does not cause preferential engulfment.
- Tighter spacing between ligand-receptor molecules (4T origami pegboard, Fig. 2B) increases the probability of engulfment initiation as well as the overall frequency of successful completion of engulfment.
- 4T ligand spacing was still preferentially engulfed when 4T, 4M, and 4S ligands were replaced with higher-affinity DNA oligos (Fig. 2C), suggesting avidity does not play a role in engulfment efficiency.
- Spatial organization of ligand-receptor molecules can affect downstream signaling events that occur in phagocytic cup formation, such as an increase in receptor phosphorylation observed in the more tightly clustered ligands. However, increasing the number of intracellular signalling modules (ITAMs) was not sufficient to surpass the threshold required to initiate phagocytic engulfment.
Why we chose this prelight
The use of DNA origami to precisely space DNA-FcɣR ligands is a fascinating approach to answer the important question of optimal FcɣR spacing and density. This controlled and tunable system yielded convincing data supporting a specific orientation of DNA-FcɣR ligands that is optimal for phagocytosis. Previous work has not offered such specific and fine control over the spacing of ligands making this system an exciting advancement for the future investigation of the cell-target interactions that drive phagocytosis and other signalling systems as well as having therapeutic implications for optimizing antibody spacing or chimeric antigen receptors.
Questions for the authors
- Does the size of the particle itself impact phagocytosis? Would the same experiment on a smaller or larger bead yield similar results?
- How does the affinity for the DNA-FcɣR ligand compare to the endogenous Fc region and how might this impact phagocytosis? Have the authors consider probing the effects of using different combinations of affinity ligands to see how weak interactions may affect phagocytosis?
- Do 4T origami pegboard spatial dynamics of ligand-receptor interactions recapitulate biologically relevant conformations such as those found on opsonized viruses or bacteria?
- According to Bakalar et al. (2018), antigen height can enhance/ impair phagocytic efficiency depending on the distance between the target cell and macrophage. We noticed that the height of the ligand remained unchanged across the experiments performed – do the authors believe that changing the height would affect which spatial arrangement (4T, 4M, 4S) or which cluster size leads to preferential engulfment?
References
Tight nanoscale clustering of Fcγ-receptors using DNA origami promotes phagocytosis
Nadja Kern, Rui Dong, Shawn M. Douglas, Ronald D. Vale, Meghan A. Morrissey
bioRxiv 2021.03.18.436011; doi: https://doi.org/10.1101/2021.03.18.436011
Dilillo, D. J., Tan, G. S., Palese, P., & Ravetch, J. V. (2014). Broadly neutralizing hemagglutinin stalk-specific antibodies require FcR interactions for protection against influenza virus in vivo. Nature Medicine, 20(2), 143–151.
Erwig, L. P., & Gow, N. A. R. (2016, February 15). Interactions of fungal pathogens with phagocytes. Nature Reviews Microbiology, Vol. 14, pp. 163–176.
Goodridge, H. S., Underhill, D. M., & Touret, N. (2012). Mechanisms of Fc Receptor and Dectin-1 Activation for Phagocytosis. Traffic, 13(8), 1062–1071.
Jaumouillé, V., Farkash, Y., Jaqaman, K., Das, R., Lowell, C. A., & Grinstein, S. (2014). Actin cytoskeleton reorganization by syk regulates fcγ receptor responsiveness by increasing its lateral mobility and clustering. Developmental Cell, 29(5), 534–546.
Nimmerjahn, F., & Ravetch, J. V. (2008). Fcγ receptors as regulators of immune responses. Nature Reviews Immunology.
Zhang, Y., Hoppe, A. D., & Swanson, J. A. (2010). Coordination of Fc receptor signaling regulates cellular commitment to phagocytosis. Proceedings of the National Academy of Sciences of the United States of America, 107(45), 19332–19337.
Posted on: 14 April 2021 , updated on: 21 April 2021
doi: Pending
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)
3 years
Meghan Morrissey
Thank you for writing the prelight, and the very thoughtful comments!
Here are some of our thoughts.
Does the size of the particle itself impact phagocytosis? Would the same experiment on a smaller or larger bead yield similar results?
-Great questions! Yes, we’ve found that smaller particles are much easier for the macrophage to engulf. However, the specificity is a little less for smaller beads – small beads with no ‘Eat me’ signal are engulfed much more than large beads with no ‘Eat me’ signals (doi.org/10.7554/eLife.36688). We didnt specifically look at whether larger beads had a stronger requirement for clustered ligands. It would be an interesting future experiment!
How does the affinity for the DNA-FcɣR ligand compare to the endogenous Fc region and how might this impact phagocytosis? Have the authors consider probing the effects of using different combinations of affinity ligands to see how weak interactions may affect phagocytosis?
-This is an excellent question as well! The FcɣR is composed of an alpha chain and two gamma chains. The gamma chain mediates intracellular signal transduction, and in mice there is only one version. However there are different alpha chains and the main difference between these alpha chains is their affinity for IgG. Why has evolution given us this system – is there an advantage to combining different affinities? We don’t know but are interested! For most experiments in the paper, we used a DNA receptor that should have a similar affinity to the medium affinity endogenous alpha chains.
Do 4T origami pegboard spatial dynamics of ligand-receptor interactions recapitulate biologically relevant conformations such as those found on opsonized viruses or bacteria?
-Hm, good question! We have thought most about how our findings compare to antibodies bound to the surface of the cell (like a therapeutic antibody, or an antibody bound to an infected cell). In that case, if the antigen is mobile, the ligated Fc Receptors will coalesce into clusters. This observation was what made us think receptor clustering might be important. For immobile ligands, like on a bacteria cell wall, it would probably vary based on what antigen the antibody detects.
According to Bakalar et al. (2018), antigen height can enhance/ impair phagocytic efficiency depending on the distance between the target cell and macrophage. We noticed that the height of the ligand remained unchanged across the experiments performed – do the authors believe that changing the height would affect which spatial arrangement (4T, 4M, 4S) or which cluster size leads to preferential engulfment?
-We agree the Bakalar et al paper is beautiful! Bakalar and others have provided very strong evidence that Fc Receptor activation requires spatial segregation of receptors and the bulky phosphatases that de-activate these receptors. The space between the macrophage and target is one factor controlling phosphatase segregation. The next step in our origami project is to figure out why clustered receptors can be more potently phosphorylated. Are clusters like the 4T better able to exclude phosphatases? Its one of our hypotheses! Looking at how tall 4Ts compare to tall 4Ss might help us answer that.