Double-strand breaks in facultative heterochromatin require specific movements and chromatin changes for efficient repair
Posted on: 15 December 2023
Preprint posted on 1 December 2023
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-024-53313-2
Histone demethylase dUtx safeguards facultative heterochromatin integrity in response to DNA breaks
Selected by Pierre CaronCategories: cell biology, genomics, molecular biology
Background
Every day our genome suffers a significant amount of damage, including DNA double-strand breaks (DSBs). This type of damage can be repaired by a few different mechanisms, distinguished by their ability to faithfully repair DNA and restore the sequence that existed prior to the damage. The choice of repair pathway for DNA double-strand breaks has an impact on the appearance of DNA mutations, which can be the onset of pathologies such as cancer. Of the various existing repair mechanisms, non-homologous end-joining (NHEJ) and homologous recombination (HR) are the ones most solicited by cells. DSB repair by NHEJ can be unfaithful and therefore lead to nucleotide loss, whereas HR is a faithful process that uses a homologous sequence as a template during repair (such as the sister chromatid) (Scully et al., 2019).
In recent decades, numerous scientific articles have described how repair pathways and their choice are regulated within different nuclear compartments and chromatin environments, such as transcribed genes (Long et al., 2021; Machour & Ayoub, 2020), nucleoli (Lesage et al., 2021), telomeres (Doksani, 2019), centromeres (Scelfo & Fachinetti, 2023) and constitutive heterochromatin (Kendek et al., 2021). However, it remains unclear how DSB repair mechanisms are regulated within facultative heterochromatin domains and what their consequences are for the integrity of these chromatin domains.
In this preprint (Marieke R. Wensveen et al., 2023), the Janssen laboratory has used Drosophila as a model to induce and study DSBs within euchromatin and facultative heterochromatin domains (polycomb bodies), characterized by the presence and enrichment of methylation of lysine 27 (lys27) of histone H3 (H3K27me3) (Figure 1A). More specifically, the authors integrated the DR-white reporter system (Figure 1B) (Janssen et al., 2016; Do et al., 2014) into euchromatin and facultative heterochromatin loci, in order to (1) monitor DSBs’ mobility within these domains, (2) measure the levels of H3 methylation specific to facultative heterochromatin (H3K27me3) and (3) identify the repair mechanisms at work within these domains. These approaches enabled the authors to reveal that DSBs induced within facultative heterochromatin are rapidly excluded from these domains to be repaired by homologous recombination (HR). Remarkably, the authors show that the movement of DSBs away from these domains requires the demethylation of lys27 of histone H3 orchestrated by the histone demethylase dUtx.
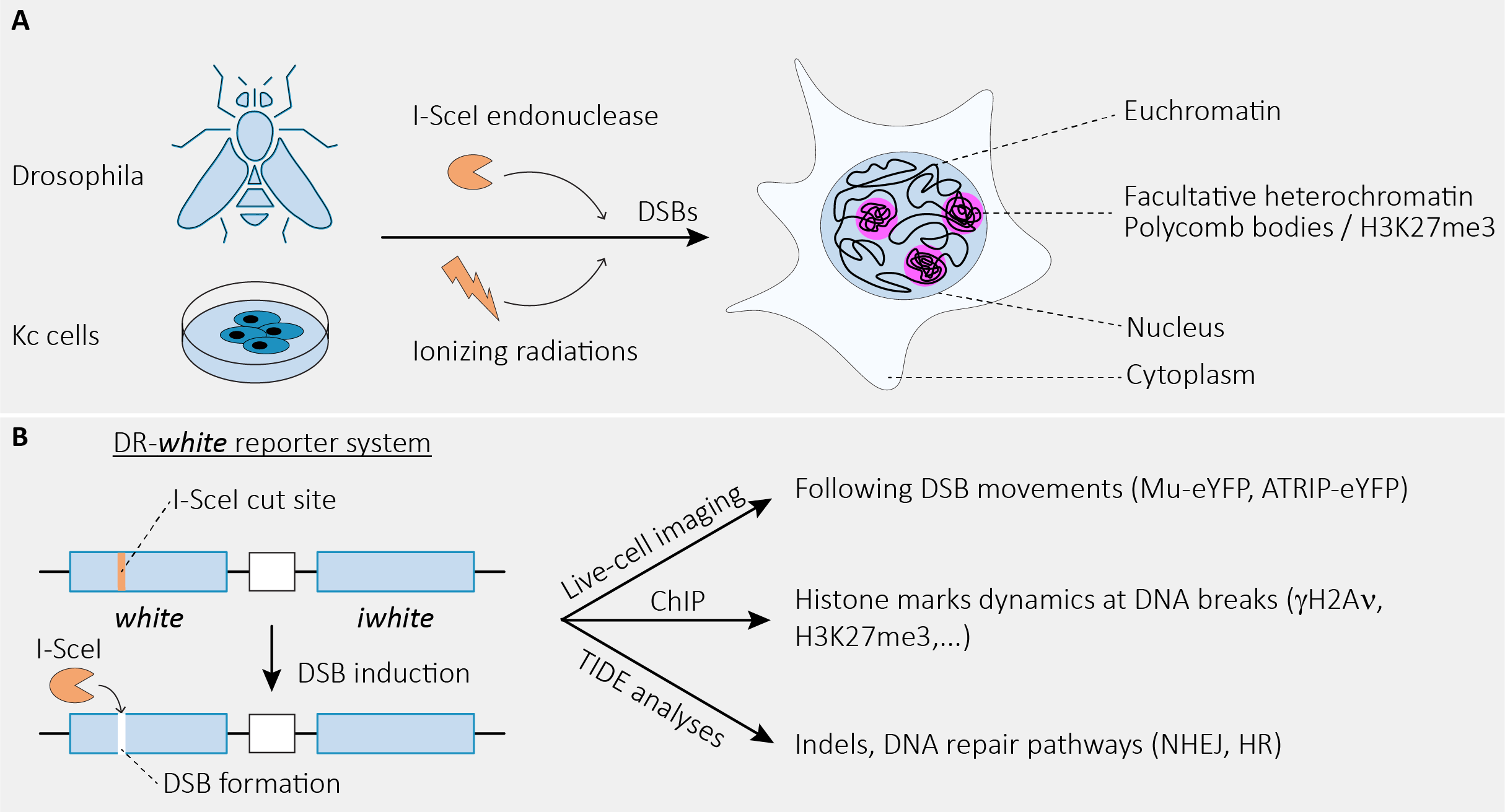
Figure 1. (A) Cartoon illustrating the models and strategies employed by the authors to study the dynamics and repair of facultative heterochromatic DSBs in Drosophila. (B) Simplified cartoon depicting the DR-white reporter system integrated in different genomic loci and its use in studying DNA repair in Drosophila tissues. (Inspired by and adapted from (Janssen et al., 2016; Marieke R. Wensveen et al., 2023))
Key findings
Studying DNA repair in facultative heterochromatin in Drosophila
With the aim to study the repair of DSBs in facultative heterochromatin in Drosophila tissues, the authors introduced the DR-white reporter system in two loci localized in euchromatin and three loci in facultative heterochromatin. As anticipated, heterochromatic insertions showed an enrichment for the histone mark H3K27me3 when compared to euchromatic insertions. Furthermore, monitoring γH2Aν levels by ChIP and immunofluorescence in larvae (third instar) and in imaginal discs, respectively, showed that I-SceI can induce DSBs in both euchromatin and facultative heterochromatin loci to the same extent. Finally, the analysis of the repair products by Sanger sequencing – followed by TIDE analyses – revealed that all DR-white insertions employed NHEJ (67-83%) and HR (17-37%) to the same extent.
DSBs induce damaged facultative heterochromatin exclusion from polycomb bodies and lead to H3K27me3 reduction
To study the dynamics of facultative heterochromatic DSBs induced by the endonuclease I-SceI or ionizing radiation, the authors performed live-cell imaging experiments during which they tracked DSB breaks using the fluorescently-tagged repair factor Mu2, which binds the histone mark γH2Aν. In a remarkable way, the vast majority of Mu2-tagged DNA breaks moved outside the polycomb bodies (facultative heterochromatin) within 5 to 10 minutes (Figure 2A). In addition, the authors found that H3K27me3 levels decrease in facultative heterochromatic breaks and that this is orchestrated by the histone demethylase dUtx (Figure 2B).
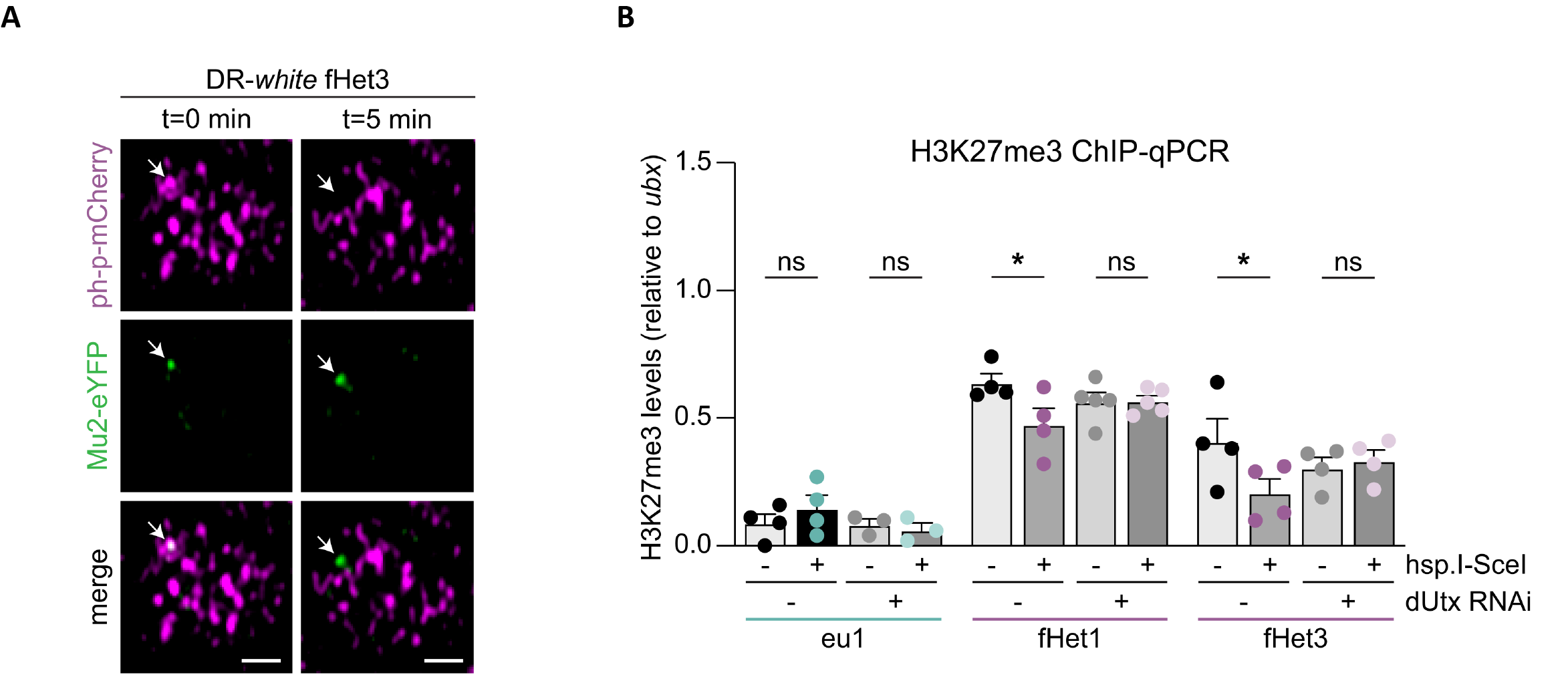
Figure 2. (figure panels from (Marieke R. Wensveen et al., 2023)). (A) Live-cell imaging in third instar larval disc to follow DNA breaks (Mu2-eYFP, green) induced within the DR-white reporter system integrated in facultative heterochromatin (polycomb bodies, ph-p-mCherry, purple). (B) ChIP-qPCR of H3K27me3 to monitor its levels in response to DSBs induced within the DR-white reporter system integrated within the euchromatin (light blue) or the facultative heterochromatin (purple), either in wild-type or dUtx-depleted cells.
dUtx-mediated H3K27me3 reduction drives facultative heterochromatic breaks movement and repair by HR
In order to assess whether the mobility of heterochromatic breaks requires chromatin changes and supports DNA repair, the author decided to follow the dynamics of the fluorescently-tagged HR repair factor ATRIP. Interestingly, ATRIP accrual to damaged chromatin occurred within the polycomb bodies. Since this factor is known to bind RPA-coated ssDNA, these findings show that early steps of HR (resection) occur within the facultative heterochromatin domains. Furthermore, while dUtx was not required for ATRIP accrual to facultative heterochromatic DNA breaks, its depletion led to reduction in DSB mobility and HR efficiency. Together these findings point to a model in which dUtx-mediated H3K27me3 reduction regulates DSB mobility outside the polycomb bodies to support DNA repair completion by HR (late steps).
Conclusion
In this preprint, the Janssen lab unveils for the first time the choreography of facultative heterochromatic DNA breaks within the nuclear space. They show that these domains employ both NHEJ and HR to repair DSBs. In addition, they reveal that the early steps of HR occur within the polycomb bodies while the later steps of this repair mechanism are orchestrated by dUTX-mediated H3K37me3 reduction which drives DNA breaks mobility and exclusion outside of these domains to support repair completion by HR. These findings shed light on a new mechanism specific to facultative heterochromatin which helps to safeguard the stability of our genome.
What I like about this preprint
This study is of great importance since it adds a missing piece to the jigsaw puzzle that aims to map the different mechanisms coordinating chromatin dynamics and DNA repair. The use of this powerful whole-animal model and the approaches described in this preprint will undoubtedly provide new insights into the understanding of genome stability within various chromatin domains and during development.
Questions for the authors
Q1: When analyzing the repair products after sequencing and TIDE analysis, did you find any signatures that might correspond to other DSB repair mechanisms, such as Poltheta-dependent alternative end-joining? If so, do you observe a higher frequency of these repair products in facultative heterochromatin compared to euchromatin, as has been observed in mammals?
Q2: As indicated in the introduction of your article, the establishment and maintenance of facultative heterochromatin through H3K27me3 is crucial to silence specific developmental genes throughout development. Since DSBs can occur at any stage of development, do you think that the loss of H3K27me3 levels could affect the silencing of certain genes and the development of the organism (in the event of DSBs within polycomb bodies)? Conversely, do you think H3K27me3 levels are restored following DSB repair completion?
References
Do, A. T., Brooks, J. T., Le Neveu, M. K., & LaRocque, J. R. (2014). Double-Strand Break Repair Assay Determine Pathway Choice and Structure of Gene Conversion Events in Drosophila melanogaster. G3, 4(3): 425–432. doi: 10.1534/g3.113.010074
Doksani. (2019). The Response to DNA Damage at Telomeric Repeats and Its Consequences for Telomere Function. Genes, 10(4), 318. https://doi.org/10.3390/genes10040318
Janssen, A., Breuer, G. A., Brinkman, E. K., van der Meulen, A. I., Borden, S. V., van Steensel, B., Bindra, R. S., LaRocque, J. R., & Karpen, G. H. (2016). A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes & Development, 30(14), 1645–1657. https://doi.org/10.1101/gad.283028.116
Kendek, A., Wensveen, M. R., & Janssen, A. (2021). The Sound of Silence: How Silenced Chromatin Orchestrates the Repair of Double-Strand Breaks. Genes, 12(9), 1415. https://doi.org/10.3390/genes12091415
Lesage, E., Clouaire, T., & Legube, G. (2021). Repair of DNA double-strand breaks in RNAPI- and RNAPII-transcribed loci. DNA Repair, 104, 103139. https://doi.org/10.1016/j.dnarep.2021.103139
Long, Q., Liu, Z., & Gullerova, M. (2021). Sweet Melody or Jazz? Transcription Around DNA Double-Strand Breaks. Frontiers in Molecular Biosciences, 8. https://doi.org/10.3389/fmolb.2021.655786
Machour, F. E., & Ayoub, N. (2020). Transcriptional Regulation at DSBs: Mechanisms and Consequences. Trends in Genetics, 36(12), 981–997. https://doi.org/10.1016/j.tig.2020.01.001
Marieke R. Wensveen, Aditya A. Dixit, Serafin U. Colmenares, & Aniek Janssen. (2023). Double-strand breaks in facultative heterochromatin require specific movements and chromatin changes for efficient repair. BioRxiv. https://doi.org/https://www.biorxiv.org/content/10.1101/2023.12.01.569547v1
Scelfo, A., & Fachinetti, D. (2023). Centromere: A Trojan horse for genome stability. DNA Repair, 130, 103569. https://doi.org/10.1016/j.dnarep.2023.103569
Scully, R., Panday, A., Elango, R., & Willis, N. A. (2019). DNA double-strand break repair-pathway choice in somatic mammalian cells. Nature Reviews Molecular Cell Biology, 20(11), 698–714. https://doi.org/10.1038/s41580-019-0152-0
Acknowledgements
I thank Dr. Reinier Prosée (Prelights team) for critical reading of the manuscript.
doi: https://doi.org/10.1242/prelights.36228
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Taxane-Induced Conformational Changes in the Microtubule Lattice Activate GEF-H1-Dependent RhoA Signaling
Vibha SINGH
PIP5K-Ras bistability triggers plasma membrane symmetry breaking to define cellular polarity and regulate migration
Vibha SINGH
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the molecular biology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the cell biology category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
December in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cell cycle and division 2) cell migration and cytoskeleton 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Matthew Davies et al. |
November in preprints – the CellBio edition
This is the first community-driven preList! A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. Categories include: 1) cancer cell biology 2) cell cycle and division 3) cell migration and cytoskeleton 4) cell organelles and organisation 5) cell signalling and mechanosensing 6) genetics/gene expression
| List by | Felipe Del Valle Batalla et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the molecular biology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)