Tunable molecular tension sensors reveal extension-based control of vinculin loading
Preprint posted on 12 February 2018 https://www.biorxiv.org/content/early/2018/02/12/264549
Article now published in eLife at http://dx.doi.org/10.7554/elife.33927
The tension sensors of the future - better optimized genetically encoded probes reveal novel mechanistic detail in how mechanotransduction works
Selected by Amanda HaageCategories: biophysics, cell biology
Why This Is Cool – It is now widely accepted that besides responding to chemical cues in their environment, cells also participate in mechanotransduction, a process by which they sense and respond to physical aspects of the environment (i.e. matrix stiffness or shear stress, etc.). The main way cells do this is through mechanosensitive signaling or the conformational change of load-bearing proteins in response to force. The main way this has been studied is through FRET-based tension sensors that are genetically encoded into those load-bearing proteins. The problem addressed by this work is that to date, the design of these tension sensors has been based on models created in cell-free systems, but they are then exclusively used in cells. Here they provide a novel and validated method for predicting tension sensor sensitivity in cells. This allows them to create an optimized tension sensor for the load-bearing protein vinculin that is determined to be 300% better by their calculations. They use this optimized sensor to demonstrate a gradient of forces across focal adhesions at the cell periphery. They are also able to use sensors of varying length to determine that protein extension, rather than the force experienced by the protein, may be the major mechanism of mechanical signaling in cells. This work provides an incredibly useful tool in mapping out the future of precise new sensor design. It demonstrates the usefulness of optimized sensors to uncover new mechanisms of mechanical signaling.
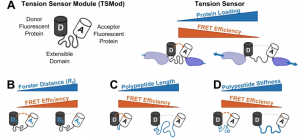
Why I Selected It – I think the model presented here will quickly be adopted as the new standard in molecular tension sensor design. They have essentially eliminated the previous system of designing that was based on literature guesswork and then checking if sensors work through extensive cellular characterization. As someone interested in mechanobiology, I look forward to seeing a new crop of tension sensors that are designed based on this model and expanded to other load-bearing proteins.
Open Questions –
- Are there any reasons the principles of tension sensor design discussed here could not be expanded to proteins other than vinculin?
- In which cellular contexts would either an extension- or force-based control mechanism be more advantageous? Why would this be a context-specific way to modulate mechanical signaling?
- Will tension sensors ever be useful in vivo?
Related Research –
- The first calibrated genetically-encoded molecular tension sensor module & the original vinculin tension sensor
- Grashoff, C., Hoffman, B. D., Brenner, M. D., Zhou, R., Parsons, M., Yang, M. T., McLean, M. A., Sligar, S. G., Chen, C. S., Ha, T., Schwartz, M. A. (2010). Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature, 466: 263-266. doi:10.1038/nature09198
- Review of the field and current limitations
- Freikamp, A., Mehlich, A., Klingner, C., Grashoff, C. (2016b). Investigating piconewton forces in cells by FRET-based molecular force microscopy. J Struct Biol. doi:10.1016/j.jsb.2016.03.011
Posted on: 4 March 2018
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Structural basis of respiratory complexes adaptation to cold temperatures
Actin polymerization drives lumen formation in a human epiblast model
Learning a conserved mechanism for early neuroectoderm morphogenesis
Also in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
preLists in the biophysics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)