Human skeletal muscle CD90+ fibro-adipogenic progenitors are associated with muscle degeneration in type 2 diabetic patients
Preprint posted on 25 August 2020 https://www.biorxiv.org/content/10.1101/2020.08.25.243907v1
Article now published in Cell Metabolism at http://dx.doi.org/10.1016/j.cmet.2021.10.001
Human skeletal muscle fibro-adipogenic remodelling and degeneration in type 2 diabetes
Selected by Osvaldo Contreras, Nicolas CollaoCategories: biochemistry, bioinformatics, cell biology, genomics, pathology, physiology
Background
Obesity and obesity-related disorders are a global epidemic (for review see Blüher, 2019). People with obesity have increased risk of morbidity and mortality mainly due to comorbidities associated with excessive weight gain, hyperglycemia and metabolic impairment (Piché et al., 2020). Therefore, obesity negatively impacts the quality of life of obese individuals and represents a major burden on our ageing society. Type 2 diabetes (T2D) is a progressive condition and the risk of developing T2D dramatically increases with obesity, insufficient physical activity and ageing. In T2D our cells become resistant to the effects of insulin (a peptide hormone which reduces the levels of glucose in the blood) and/or the pancreas loses its competence to produce insulin, which results in hyperglycemia. Additionally, obese individuals with T2D develop muscle atrophy – a condition where muscles are smaller than normal – and fibro-fatty infiltration, both of which negatively affect muscle contractile function (Hilton et al., 2008; Tam et al., 2014).
The adult mammalian skeletal muscles are composed of many cell types, and therefore their niche composition is highly complex and dynamic, especially upon damage. Muscle cells communicate by physically interacting and/or secreting a plethora of factors to maintain muscle homeostasis. When muscle homeostasis is acutely disrupted following injury, adult muscle stem cells (MuSCs) (also known as satellite cells) activate, proliferate, self-renew, and differentiate to fuse with pre-existent myofibers to effectively restore the lost muscle to its previous condition, a phenomenon called adult skeletal muscle regeneration (Joe et al., 2010). However, during chronic degenerative conditions, such as obesity, T2D, and ageing, the muscle milieu is gradually disrupted, leading to a pathophysiological condition that triggers exacerbated accumulation of extracellular matrix (ECM) and adipose tissue. This associates with muscle metabolic profile dysregulation, increased tissue stiffness and reduced contraction of the affected muscles (Buras et al., 2019; Collao et al., 2020; Teng et al., 2019). Fibro-adipogenic progenitors (FAPs) are the fibroblasts of mammalian muscles with multipotency towards all the mesenchymal cell lineages (Contreras et al., 2019; Eisner et al., 2020; Kopinke et al., 2017; Scott et al., 2019; Uezumi et al., 2010, 2014). FAPs are diverse and dynamic cells required for adult muscle maintenance and effective muscle regeneration (De Micheli et al., 2020; Giordani et al., 2019; Malecova et al., 2018; Marinkovic et al., 2019; Wosczyna et al., 2019). However, FAPs can contribute to muscle pathology, where chronic injury and inflammation blunt muscle regeneration and lead to progressive tissue degeneration (Contreras et al., 2016; Hogarth et al., 2019; Reggio et al., 2020). Although important progress has been made in understanding the heterogeneity of the stromal compartment in muscle health, regeneration, and disease (Oprescu et al., 2020; Rubenstein et al., 2020; Scott et al., 2019), the cellular and molecular responses of human skeletal muscle FAPs to gradual degenerative diseases are underexplored.
In this preprint, Farup and colleagues unveiled a subpopulation of human muscle-resident FAPs that associates with progressive muscle degeneration in T2D patients. They suggested that a subset of muscle FAPs (Lin–CD56–CD82–CD34+CD90+) associates with enhanced fibrosis and ectopic adipose tissue in degenerative T2D settings, shedding new light on the role of fibro-adipogenic progenitors underpinning skeletal muscle degenerative fibro-fatty remodelling in the obese and T2D population.
Obese individuals with T2D develop muscle atrophy – a condition where muscles are smaller than normal – and fibro-fatty infiltration, both of which negatively affect muscle contractile function.
Key findings
First, using skeletal muscle biopsies, the authors profiled bulk transcriptomic changes from 3 separate groups: individuals with obesity, T2D, or insulin-treated T2D (itT2D), which represented a disease progression-type model that associates with the severity of insulin resistance (obese<T2D<itT2D). Farup et al. found increased ECM remodelling gene signatures in itT2D, and therefore, hypothesised that FAPs influence muscle pathology in hyperglycemic patients. To further test this hypothesis, the authors isolated and characterized human FAPs from muscle biopsies based on CD90 cell-surface protein expression using fluorescent-activated cell sorting (FACS). This because neither of the tested antibodies against PDGFRA (also known as CD140a) worked -even when PDGFRA is highly expressed in human FAPs- nor Sca-1/Ly6a antigen is expressed in human cells, which are two well-characterized FAP markers in mouse skeletal muscles. The authors also corroborated that human FAPs display clonal expansion (4% for FAPs compared to 17% for MuSCs) and are distinct to other muscle-resident populations of mononuclear cells, which confirm previous findings in mice. Platelet-derived growth factors (PDGFs) bind to PDGFRα and PDGFRβ to regulate key molecular and cellular processes including proliferation, migration and gene expression. Furthermore, PDGF signaling regulates the fate of skeletal muscle FAPs (Contreras et al., 2019; Mueller et al., 2016). Farup et al. described that PDGFRA expression correlates with COLLAGEN 1A1 expression, as previously proposed (Contreras et al., 2019). Owing to this relationship, they asked whether PDGF signalling could impact the fate of FAPs. PDGF-AA treatment increased the expression of fibrillar collagen in FAPs, whereas it reduced their adipogenic differentiation. The authors showed that the PDGF-AA-mediated fibrogenic activation of FAPs associates with a metabolic switch that favours an enhanced consumption of glucose in these cells compared to non-treated cells. This increased glycolytic flux present in TGF-b-induced fibrogenic conditions seems to be required for enhanced ECM synthesis and deposition by FAPs.
The expression of the cell-surface protein CD90 has been widely used to identify tissue-resident fibroblasts and/or mesenchymal stromal/stem cells in different tissues and organs. In this preprint, combining FACS with single-cell RNA-seq, the authors showed that CD90 expression was restricted to a particular FAPs subpopulation. Remarkably, CD90 positive (CD90+) FAPs and CD90 negative (CD90–) FAPs exhibited distinct phenotypic and molecular signatures. CD90+ FAPs were bigger in size, proliferated faster, and displayed higher expression of extracellular matrix genes compared to CD90- FAPs. Glycolytic flux and maximal oxygen consumption were also higher in CD90+ FAPs compared to CD90- FAPs. These results suggest that at least two distinct FAP subpopulations are present in skeletal muscle and are distinguished by the expression of CD90/THY1. Since evidence of fibrosis was detected in skeletal muscle biopsies from patients with T2D, the authors compared the content of the CD90+, pro-fibrotic FAP subpopulation between patients with T2D and healthy controls. They found that the CD90+ FAP subpopulation was higher in muscles of individuals with T2D compared to non-T2D controls. Further, CD90+ FAPs from patients with T2D proliferated faster and had higher expression of COLLAGEN 1 compared to muscleCD90+ FAPs isolated from non-diabetic individuals. And lastly, the authors sought to pharmacologically target FAPs to prevent their excessive accumulation and fibro-fatty infiltration in muscles of T2D patients. Metformin is a usual first-line pharmacological approach to counteract T2D. Metformin treatment reduced the proliferation, oxygen consumption, and adipogenic differentiation of CD90+ FAPs but increased the glycolytic flux of these cells, suggesting a novel therapeutically targetable cellular mechanism to reduce intra/intermuscular adipose tissue deposition in T2D. Taken together, these findings describe two distinct FAP subsets in human skeletal muscles that participate in modulating the fibro-fatty infiltration of muscles in T2D, a highly prevalent condition in western countries.
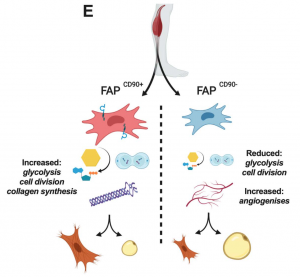
Figure. Two major FAP populations described by Farup et al., 2020. CD90 positive (CD90+) FAPs and CD90 negative (CD90–) FAPs exhibited distinct phenotypic and molecular signatures. These phenotypic differences shed new light on the role of these intriguing cells in modulating muscle degenerative fibro-fatty remodelling in the obese and T2D population.
What we liked of this preprint
The highlight of this preprint is the study of FAP heterogeneity in human muscle and how this diversity might impact skeletal muscle homeostasis. Since most of our understanding of tissue-resident FAPs comes from murine models, this study fills an important gap in our knowledge related to the role of FAPs in human degenerative disorders. Therefore, novel therapies targeting FAPs represent a promising strategy for preventing, testing and treating muscle degeneration in chronic metabolic disorders.
Future directions and questions to the authors
- The authors described that CD90 discriminates at least two subpopulations of FAPs. How is the expression of CD90 regulated in FAPs following injury and ultimately what is the role of CD90 in controlling FAP fate?
- The work is lacking scRNA-seq data sets from obese, T2D and insulin-resistant T2D patients. Perhaps this should help to understand the dynamics and heterogeneity of FAPs in progressive human chronic pathologies and to unveil the molecular and cellular responses of FAPs to weight gain and/or T2D. Are you thinking to perform these experiments?
- From what population of endogenous FAPs are CD90+ expressing FAPs descending from. Are you planning to do lineage tracing or fate-mapping experiments (in the mouse) to clarify the hierarchy and clonality of this CD90+ subpopulation of cells and their lineage origin?
- It would be interesting to know the factors that participate in the metabolic changes of muscle FAPs associated with T2D and insulin resistance over time.
- Since FAPs from healthy muscles support muscle stem cell-dependent regeneration through the secretion of trophic signals, how would be the cross-talk between CD90+ FAP and MuSCs affected in T2D? And finally, what is the behavior of MuSCs in the degenerative microenvironment of T2D patients?
- Does insulin downregulate the expression of PDGFRA? Or does the reduction of PDGFRA you observed in insulin-treated T2D patients compared to non-treated T2D patients secondary to the improved metabolic phenotype in the insulin-treated T2D individuals? What muscle-resident cells express the insulin receptor at the single-cell level? Can FAPs become resistant to insulin?
Acknowledgements
The authors are grateful to Dr Michael De Lisio (University of Ottawa) for proofreading the highlight and Dr Mate Pálfy for helpful suggestions.
References
Skeletal muscle fibrogenic and adipogenic remodelling in obesity and metabolic disorders:
- Buras, E. D., Converso-Baran, K., Davis, C. S., Akama, T., Hikage, F., Michele, D. E., … Chun, T.-H. (2019). Fibro-Adipogenic Remodeling of the Diaphragm in Obesity-Associated Respiratory Dysfunction. Diabetes, 68(1), 45–56. https://doi.org/10.2337/db18-0209
- Blüher, M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol 15, 288–298 (2019). https://doi.org/10.1038/s41574-019-0176-8
- Collao N, Farup J, De Lisio M. Role of Metabolic Stress and Exercise in Regulating Fibro/Adipogenic Progenitors. Front Cell Dev Biol. 2020;8:9. Published 2020 Jan 28. doi:10.3389/fcell.2020.00009
- Hilton, T. N., Tuttle, L. J., Bohnert, K. L., Mueller, M. J., & Sinacore, D. R. (2008). Excessive Adipose Tissue Infiltration in Skeletal Muscle in Individuals With Obesity, Diabetes Mellitus, and Peripheral Neuropathy: Association With Performance and Function. Physical Therapy, 88(11), 1336–1344. https://doi.org/10.2522/ptj.20080079
- Piché ME, Tchernof A, Després JP. Obesity Phenotypes, Diabetes, and Cardiovascular Diseases [published correction appears in Circ Res. 2020 Jul 17;127(3):e107]. Circ Res. 2020;126(11):1477-1500. doi:10.1161/CIRCRESAHA.120.316101
- Tam, C. S., Covington, J. D., Bajpeyi, S., Tchoukalova, Y., Burk, D., Johannsen, D. L., … Ravussin, E. (2014). Weight Gain Reveals Dramatic Increases in Skeletal Muscle Extracellular Matrix Remodeling. The Journal of Clinical Endocrinology & Metabolism, 99(5), 1749–1757. https://doi.org/10.1210/jc.2013-4381
- Teng S, Huang P. The effect of type 2 diabetes mellitus and obesity on muscle progenitor cell function. Stem Cell Res Ther. 2019;10(1):103. Published 2019 Mar 21. doi:10.1186/s13287-019-1186-0
FAPs and their lineage in muscle health, regeneration and disease:
- Contreras O, Rebolledo DL, Oyarzún JE, Olguín HC, Brandan E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell Tissue Res. 2016;364(3):647-660. doi:10.1007/s00441-015-2343-0
- Eisner, C., Cummings, M., Johnston, G., Tung, L.W., Groppa, E., Chang, C. and Rossi, F.M. (2020), Murine Tissue‐Resident PDGFRα+ Fibro‐Adipogenic Progenitors Spontaneously Acquire Osteogenic Phenotype in an Altered Inflammatory Environment. J Bone Miner Res, 35: 1525-1534. doi:10.1002/jbmr.4020
- Hogarth, M.W., Defour, A., Lazarski, C., Gallardo, E., Diaz Manera, J., Partridge, T.A., Nagaraju, K., Jaiswal, J.K. (2019). Fibroadipogenic progenitors are responsible for muscle loss in limb girdle muscular dystrophy 2B. Nat Commun. 3;10(1):2430. doi: 10.1038/s41467-019-10438-z.
- Joe AW, Yi L, Natarajan A, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12(2):153-163. doi:10.1038/ncb2015
- Kopinke, D., Roberson, E. C. and Reiter, J. F. (2017). Ciliary hedgehog signaling restricts injury-induced adipogenesis. Cell 170, 340-351.e312. doi:10.1016/j.cell.2017.06.035
- Mueller AA, van Velthoven CT, Fukumoto KD, Cheung TH, Rando TA. Intronic polyadenylation of PDGFRalpha in resident stem cells attenuates muscle fibrosis. Nature. 2016; 540:276–279
- Reggio A, Rosina M, Krahmer N, et al. Metabolic reprogramming of fibro/adipogenic progenitors facilitates muscle regeneration. Life Sci Alliance. 2020;3(3):e202000646. Published 2020 Feb 4. doi:10.26508/lsa.202000660
- Uezumi A, Fukada S, Yamamoto N, et al. Identification and characterization of PDGFRα+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis. 2014;5(4):e1186. Published 2014 Apr 17. doi:10.1038/cddis.2014.161
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12(2):143-152. doi:10.1038/ncb2014
- Wosczyna MN, Konishi CT, Perez Carbajal EE, et al. Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep. 2019;27(7):2029-2035.e5. doi:10.1016/j.celrep.2019.04.074
Skeletal muscle FAP diversity:
- Contreras O, Cruz-Soca M, Theret M, et al. Cross-talk between TGF-β and PDGFRα signaling pathways regulates the fate of stromal fibro-adipogenic progenitors. J Cell Sci. 2019;132(19):jcs232157. Published 2019 Oct 9. doi:10.1242/jcs.232157
- De Micheli AJ, Laurilliard EJ, Heinke CL, et al. Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Rep. 2020;30(10):3583-3595.e5. doi:10.1016/j.celrep.2020.02.067
- Giordani L, He GJ, Negroni E, et al. High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol Cell. 2019;74(3):609-621.e6. doi:10.1016/j.molcel.2019.02.026
- Malecova B, Gatto S, Etxaniz U, et al. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat Commun. 2018;9(1):3670. Published 2018 Sep 10. doi:10.1038/s41467-018-06068-6
- Marinkovic M, Fuoco C, Sacco F, et al. Fibro-adipogenic progenitors of dystrophic mice are insensitive to NOTCH regulation of adipogenesis. Life Sci Alliance. 2019;2(3):e201900437. Published 2019 Jun 25. doi:10.26508/lsa.201900437
- Oprescu SN, Yue F, Qiu J, Brito LF, Kuang S. Temporal Dynamics and Heterogeneity of Cell Populations during Skeletal Muscle Regeneration. iScience. 2020;23(4):100993. doi:10.1016/j.isci.2020.100993
- Rubenstein AB, Smith GR, Raue U, et al. Single-cell transcriptional profiles in human skeletal muscle. Sci Rep. 2020;10(1):229. Published 2020 Jan 14. doi:10.1038/s41598-019-57110-6
- Scott RW, Arostegui M, Schweitzer R, Rossi FMV, Underhill TM. Hic1 Defines Quiescent Mesenchymal Progenitor Subpopulations with Distinct Functions and Fates in Skeletal Muscle Regeneration. Cell Stem Cell. 2019; 25(6):797-813.e9. doi:10.1016/j.stem.2019.11.004
Posted on: 10 September 2020 , updated on: 16 September 2020
doi: https://doi.org/10.1242/prelights.24354
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the bioinformatics category:
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Holimap: an accurate and efficient method for solving stochastic gene network dynamics
Also in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the genomics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
Also in the pathology category:
Hypoxia blunts angiogenic signaling and upregulates the antioxidant system in elephant seal endothelial cells
H2O2 sulfenylates CHE linking local infection to establishment of systemic acquired resistance
Bacterial filamentation is an in vivo mechanism for cell-to-cell spreading
Also in the physiology category:
Unlocking the secrets of kangaroo locomotor energetics: Postural adaptations underpin increased tendon stress in hopping kangaroos
Changes in surface temperatures reveal the thermal challenge associated with catastrophic moult in captive Gentoo penguins
Torpor energetics are related to the interaction between body mass and climate in bats of the family Vespertilionidae
preLists in the biochemistry category:
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the bioinformatics category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the cell biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the genomics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the pathology category:
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
Also in the physiology category:
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |











 (1 votes)
(1 votes)